Iodine »
PDB 1lkr-1pvh »
1mqg »
Iodine in PDB 1mqg: Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution
Protein crystallography data
The structure of Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution, PDB code: 1mqg
was solved by
R.Jin,
T.G.Banke,
M.L.Mayer,
S.F.Traynelis,
E.Gouaux,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.71 / 2.15 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.006, 88.716, 58.907, 90.00, 99.24, 90.00 |
| R / Rfree (%) | 18.5 / 23.3 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution
(pdb code 1mqg). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 3 binding sites of Iodine where determined in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution, PDB code: 1mqg:
Jump to Iodine binding site number: 1; 2; 3;
In total 3 binding sites of Iodine where determined in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution, PDB code: 1mqg:
Jump to Iodine binding site number: 1; 2; 3;
Iodine binding site 1 out of 3 in 1mqg
Go back to
Iodine binding site 1 out
of 3 in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution
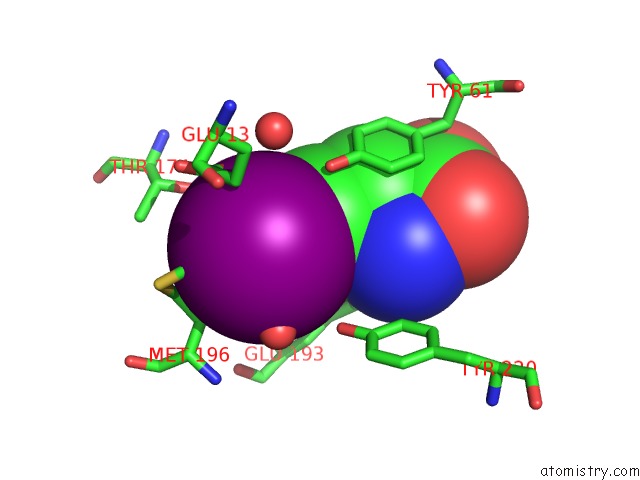
Mono view
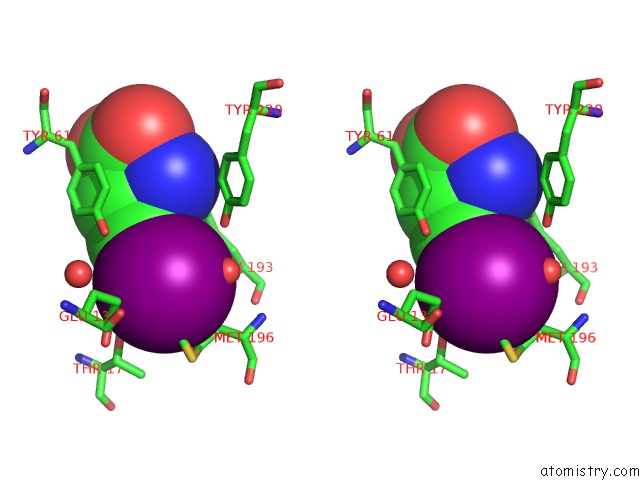
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution within 5.0Å range:
|
Iodine binding site 2 out of 3 in 1mqg
Go back to
Iodine binding site 2 out
of 3 in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution
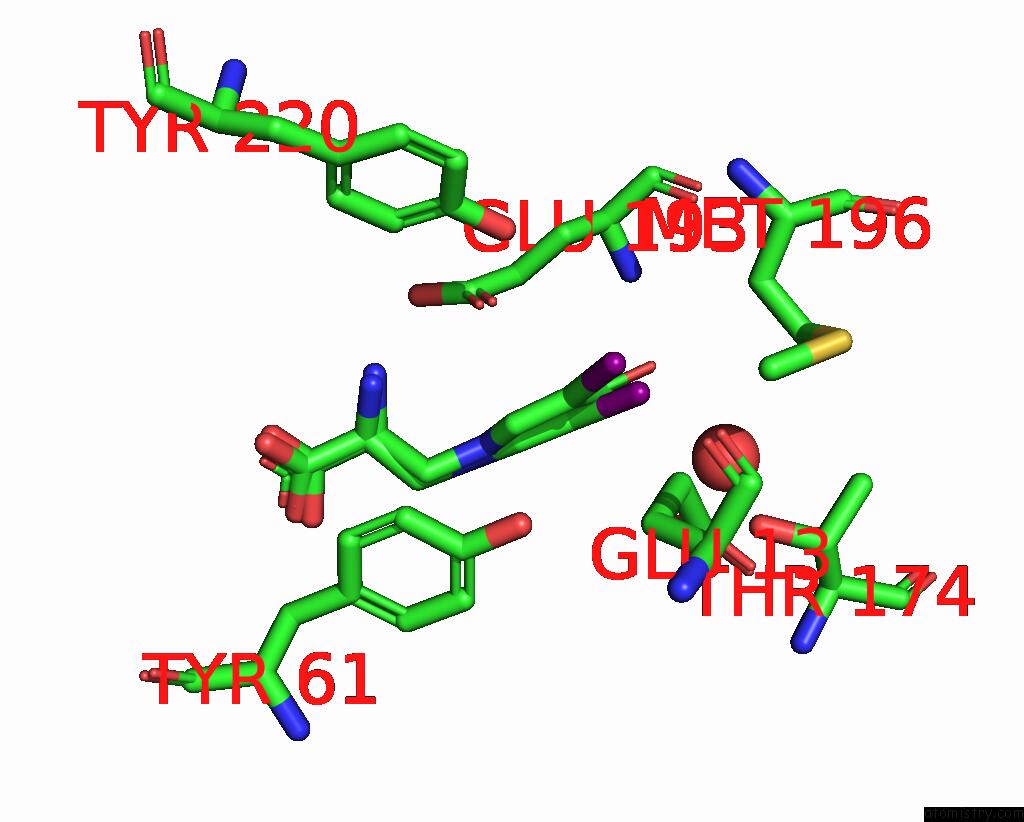
Mono view
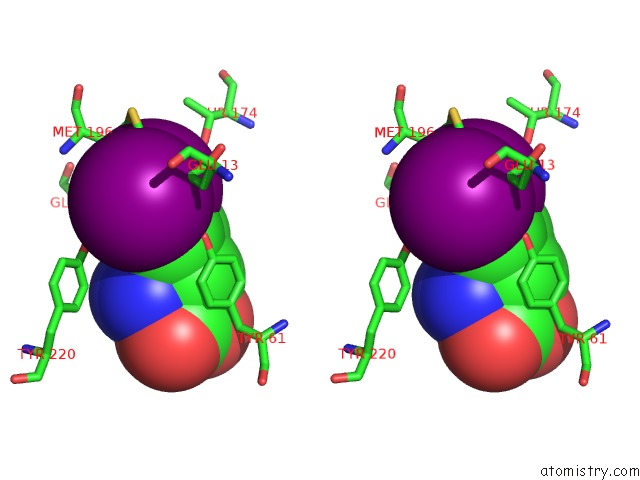
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution within 5.0Å range:
|
Iodine binding site 3 out of 3 in 1mqg
Go back to
Iodine binding site 3 out
of 3 in the Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution
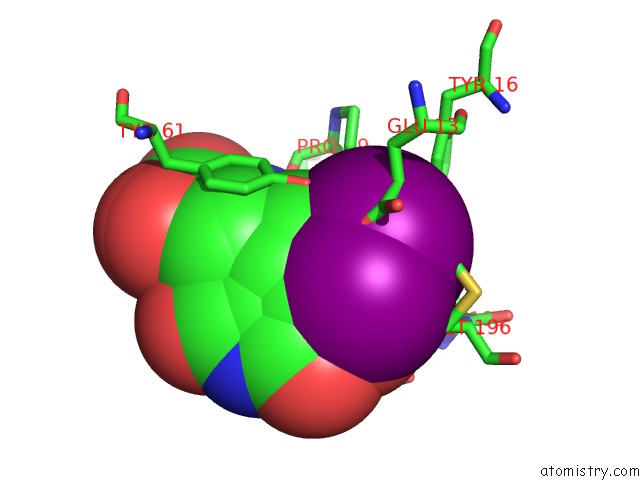
Mono view
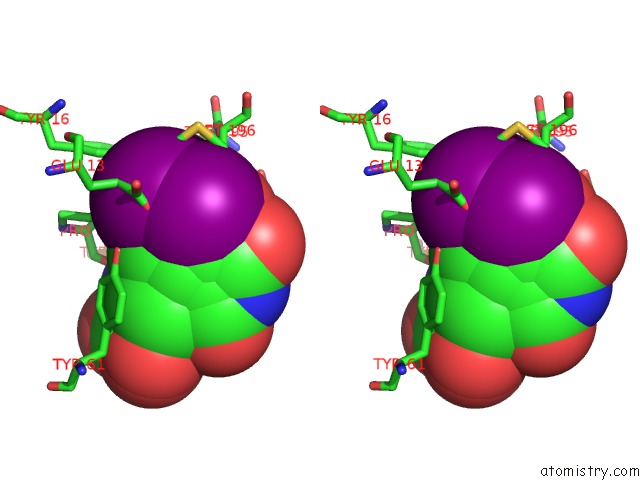
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution within 5.0Å range:
|
Reference:
R.Jin,
T.G.Banke,
M.L.Mayer,
S.F.Traynelis,
E.Gouaux.
Structural Basis For Partial Agonist Action at Ionotropic Glutamate Receptors Nat.Neurosci. V. 6 803 2003.
ISSN: ISSN 1097-6256
PubMed: 12872125
DOI: 10.1038/NN1091
Page generated: Sun Aug 11 12:28:47 2024
ISSN: ISSN 1097-6256
PubMed: 12872125
DOI: 10.1038/NN1091
Last articles
Ca in 2VZRCa in 2VZQ
Ca in 2VZP
Ca in 2VYP
Ca in 2VYO
Ca in 2VZ1
Ca in 2VY0
Ca in 2VVF
Ca in 2VWO
Ca in 2VWN