Iodine »
PDB 3usl-3zzz »
3vv6 »
Iodine in PDB 3vv6: Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
Enzymatic activity of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
All present enzymatic activity of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One:
3.4.23.46;
3.4.23.46;
Protein crystallography data
The structure of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One, PDB code: 3vv6
was solved by
S.Yonezawa,
T.Yamamoto,
H.Yamakawa,
C.Muto,
M.Hosono,
K.Hattori,
K.Higashino,
M.Sakagami,
H.Togame,
Y.Tanaka,
T.Nakano,
H.Takemoto,
M.Arisawa,
S.Shuto,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.05 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.491, 102.491, 170.156, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 24.9 / 28 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
(pdb code 3vv6). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 5 binding sites of Iodine where determined in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One, PDB code: 3vv6:
Jump to Iodine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Iodine where determined in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One, PDB code: 3vv6:
Jump to Iodine binding site number: 1; 2; 3; 4; 5;
Iodine binding site 1 out of 5 in 3vv6
Go back to
Iodine binding site 1 out
of 5 in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
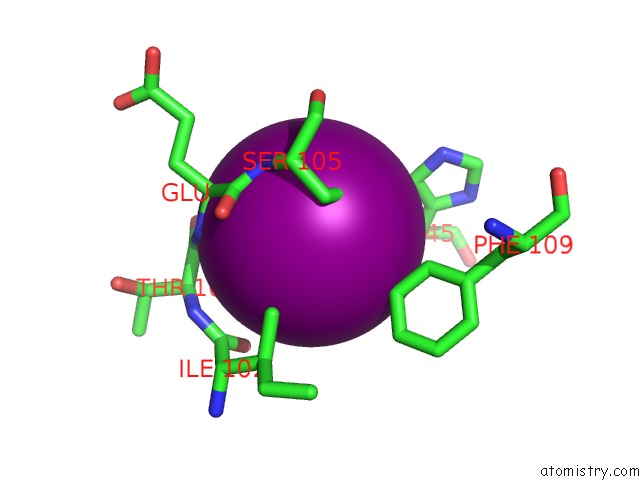
Mono view
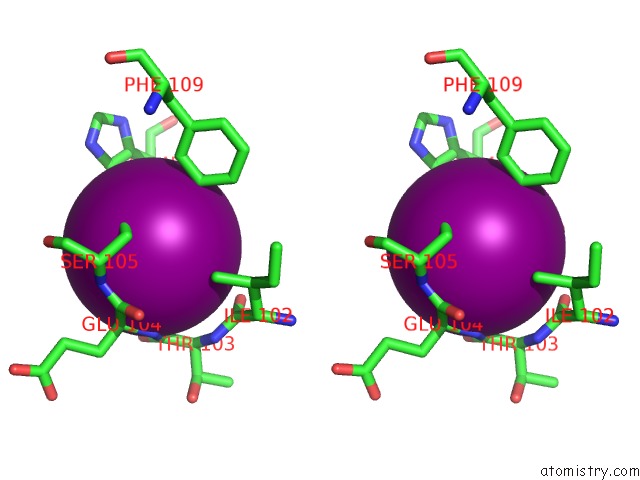
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One within 5.0Å range:
|
Iodine binding site 2 out of 5 in 3vv6
Go back to
Iodine binding site 2 out
of 5 in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
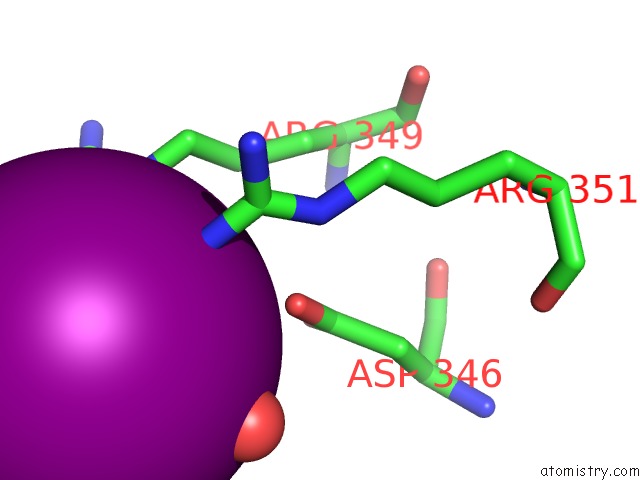
Mono view
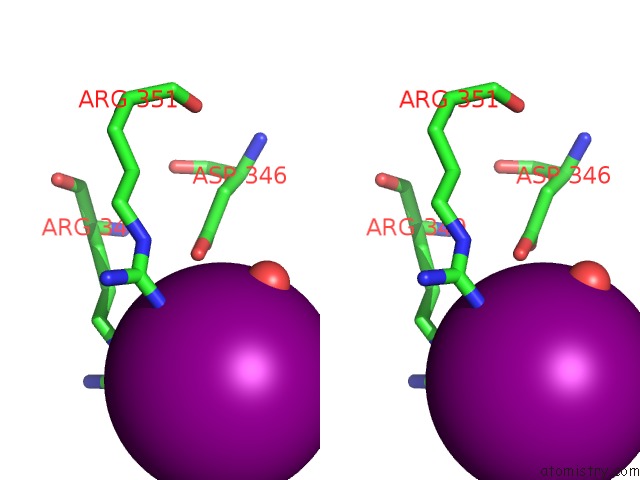
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One within 5.0Å range:
|
Iodine binding site 3 out of 5 in 3vv6
Go back to
Iodine binding site 3 out
of 5 in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
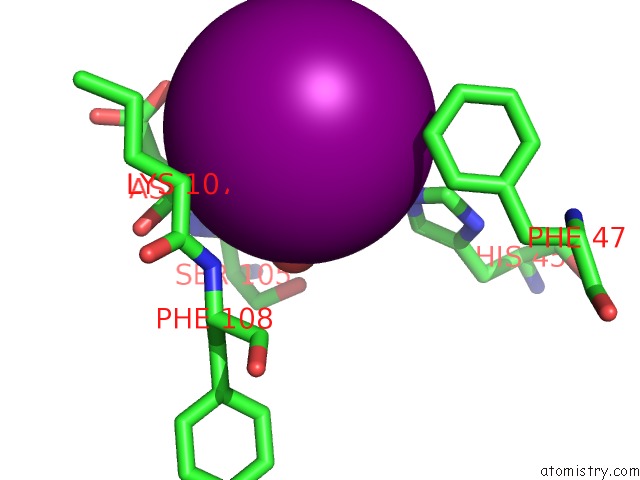
Mono view
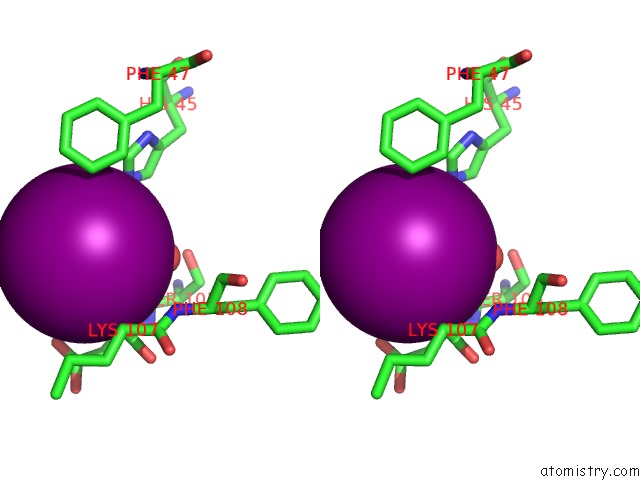
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One within 5.0Å range:
|
Iodine binding site 4 out of 5 in 3vv6
Go back to
Iodine binding site 4 out
of 5 in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
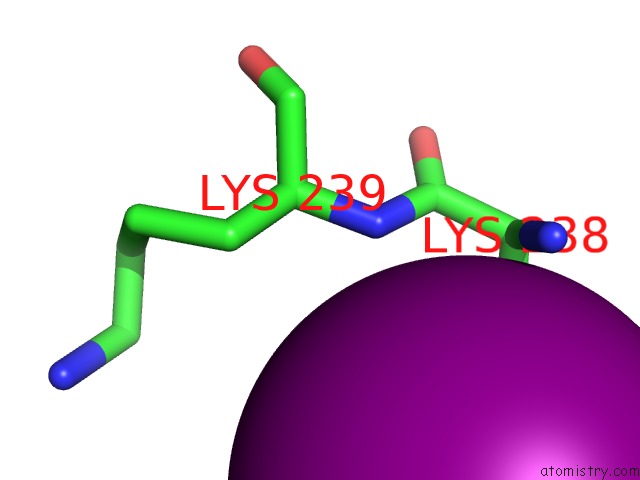
Mono view
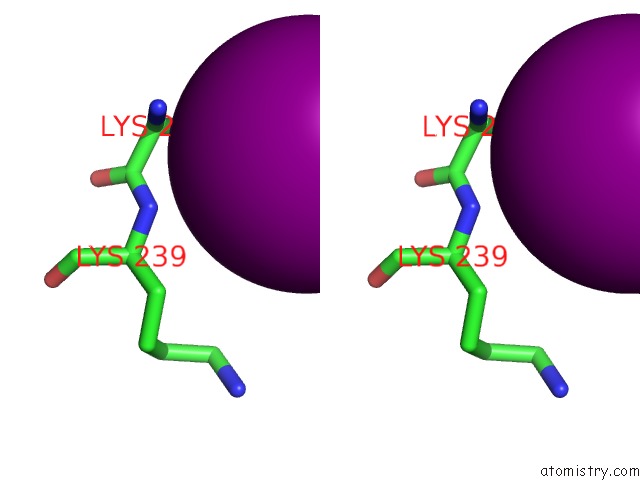
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One within 5.0Å range:
|
Iodine binding site 5 out of 5 in 3vv6
Go back to
Iodine binding site 5 out
of 5 in the Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One
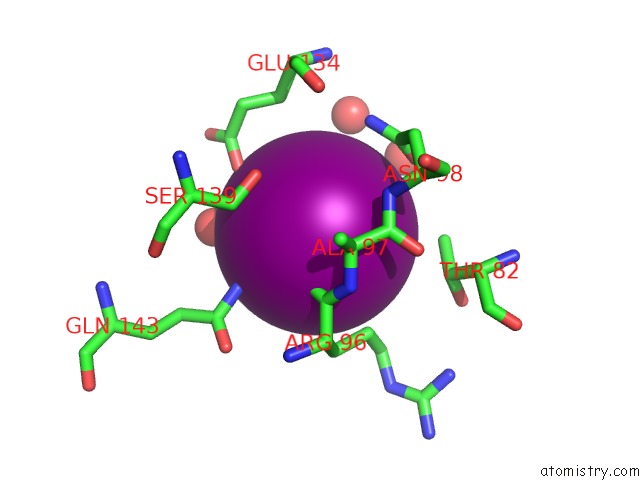
Mono view
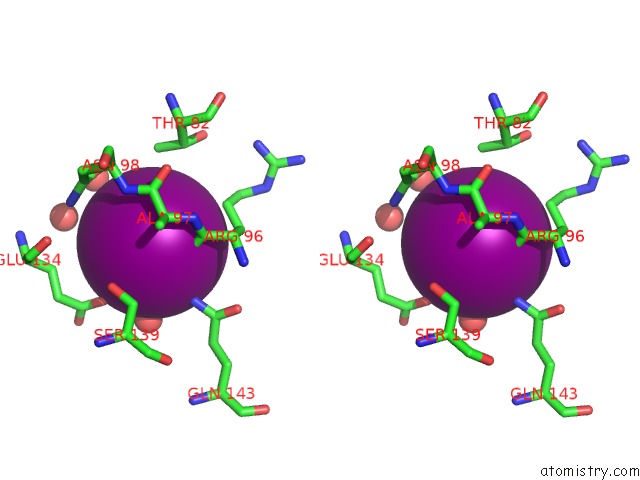
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 5 of Crystal Structure of Beta Secetase in Complex with 2-Amino-3-Methyl-6- ((1S, 2R)-2-Phenylcyclopropyl)Pyrimidin-4(3H)-One within 5.0Å range:
|
Reference:
S.Yonezawa,
T.Yamamoto,
H.Yamakawa,
C.Muto,
M.Hosono,
K.Hattori,
K.Higashino,
T.Yutsudo,
H.Iwamoto,
Y.Kondo,
M.Sakagami,
H.Togame,
Y.Tanaka,
T.Nakano,
H.Takemoto,
M.Arisawa,
S.Shuto.
Conformational Restriction Approach to Beta-Secretase (BACE1) Inhibitors: Effect of A Cyclopropane Ring to Induce An Alternative Binding Mode J.Med.Chem. V. 55 8838 2012.
ISSN: ISSN 0022-2623
PubMed: 22998419
DOI: 10.1021/JM3011405
Page generated: Fri Aug 8 15:48:35 2025
ISSN: ISSN 0022-2623
PubMed: 22998419
DOI: 10.1021/JM3011405
Last articles
K in 8GABK in 8G7M
K in 8G6X
K in 8G2N
K in 8G20
K in 8FS1
K in 8FS2
K in 8G1Z
K in 8FZ7
K in 8FZU