Iodine »
PDB 3usl-3zzz »
3zzz »
Iodine in PDB 3zzz: Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
Protein crystallography data
The structure of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2, PDB code: 3zzz
was solved by
A.Joshi,
O.Kotik-Kogan,
S.Curry,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.07 / 1.55 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.480, 60.380, 61.060, 90.00, 107.86, 90.00 |
| R / Rfree (%) | 21.7 / 22.4 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
(pdb code 3zzz). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 5 binding sites of Iodine where determined in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2, PDB code: 3zzz:
Jump to Iodine binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Iodine where determined in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2, PDB code: 3zzz:
Jump to Iodine binding site number: 1; 2; 3; 4; 5;
Iodine binding site 1 out of 5 in 3zzz
Go back to
Iodine binding site 1 out
of 5 in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
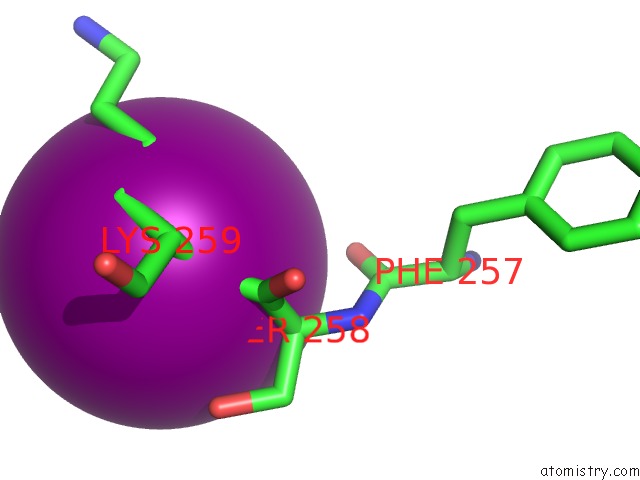
Mono view
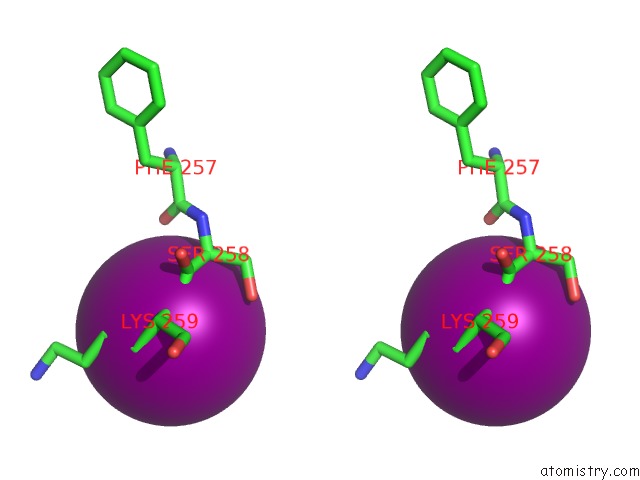
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2 within 5.0Å range:
|
Iodine binding site 2 out of 5 in 3zzz
Go back to
Iodine binding site 2 out
of 5 in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
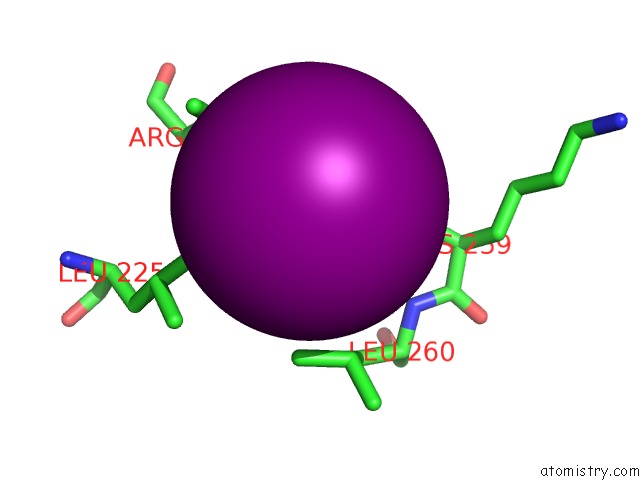
Mono view
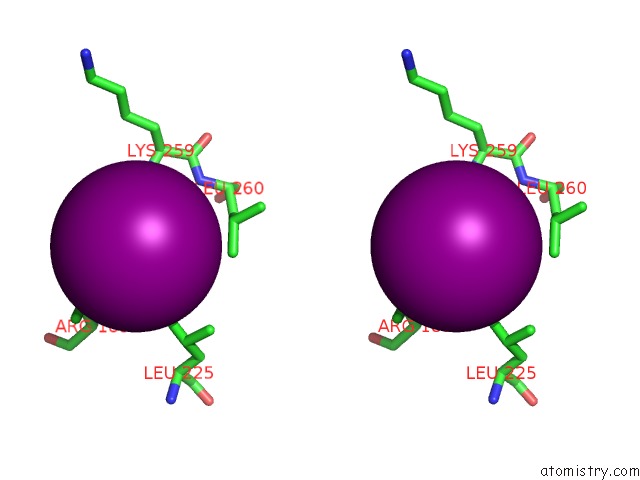
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2 within 5.0Å range:
|
Iodine binding site 3 out of 5 in 3zzz
Go back to
Iodine binding site 3 out
of 5 in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
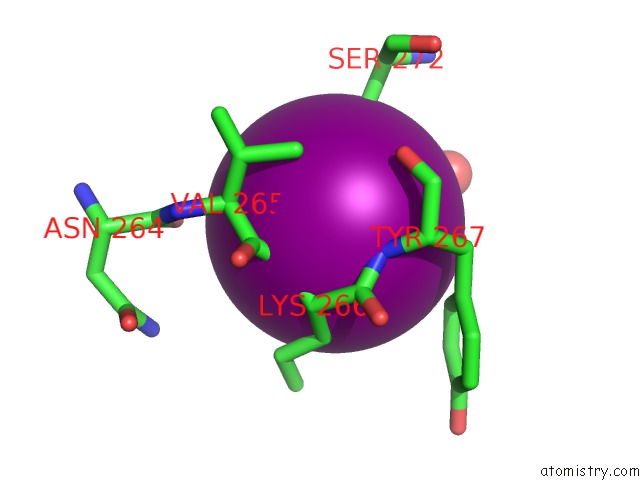
Mono view
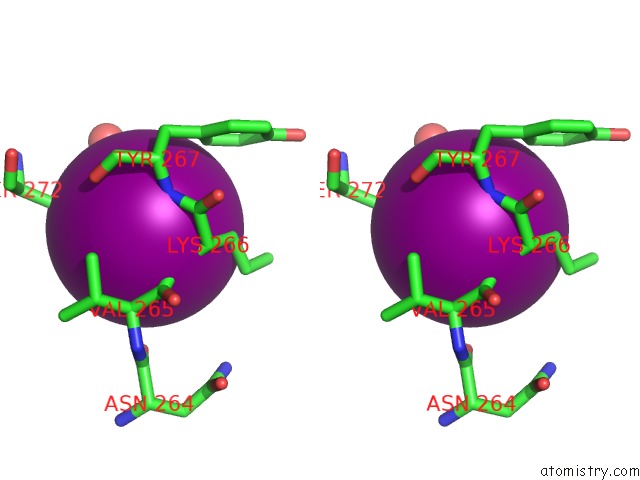
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2 within 5.0Å range:
|
Iodine binding site 4 out of 5 in 3zzz
Go back to
Iodine binding site 4 out
of 5 in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2
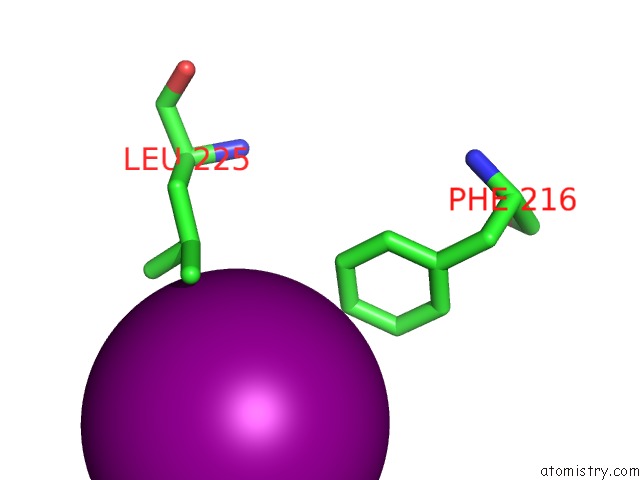
Mono view
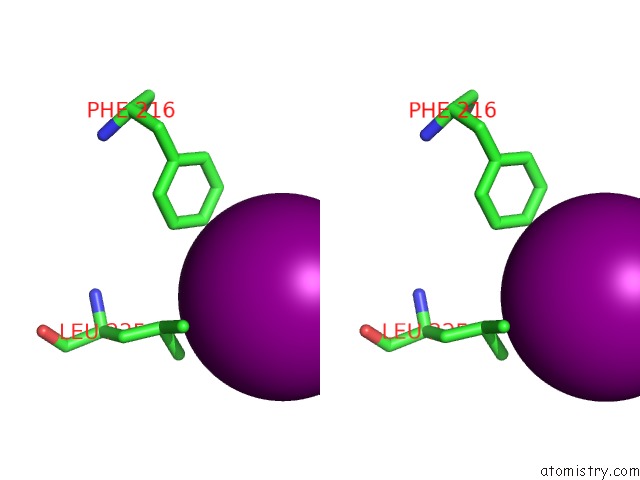
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2 within 5.0Å range:
|
Iodine binding site 5 out of 5 in 3zzz
Go back to
Iodine binding site 5 out
of 5 in the Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 5 of Crystal Structure of A RAVER1 PRI4 Peptide in Complex with Polypyrimidine Tract Binding Protein RRM2 within 5.0Å range:
|
Reference:
A.Joshi,
M.B.Coelho,
O.Kotik-Kogan,
P.J.Simpson,
S.J.Matthews,
C.W.Smith,
S.Curry.
Crystallographic Analysis of Polypyrimidine Tract-Binding Protein-RAVER1 Interactions Involved in Regulation of Alternative Splicing. Structure V. 19 1816 2011.
ISSN: ISSN 0969-2126
PubMed: 22153504
DOI: 10.1016/J.STR.2011.09.020
Page generated: Sun Aug 11 17:15:23 2024
ISSN: ISSN 0969-2126
PubMed: 22153504
DOI: 10.1016/J.STR.2011.09.020
Last articles
Cl in 5UAXCl in 5UB3
Cl in 5UBA
Cl in 5UBG
Cl in 5UB4
Cl in 5UB9
Cl in 5UAM
Cl in 5UAS
Cl in 5UAO
Cl in 5UAD