Iodine »
PDB 4p9t-4ttc »
4tjv »
Iodine in PDB 4tjv: Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
Protein crystallography data
The structure of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1, PDB code: 4tjv
was solved by
F.Luo,
Y.H.Fong,
L.W.Jiang,
K.B.Wong,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.78 / 1.65 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 32.600, 62.680, 35.810, 90.00, 109.39, 90.00 |
| R / Rfree (%) | 16.6 / 19 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
(pdb code 4tjv). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 6 binding sites of Iodine where determined in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1, PDB code: 4tjv:
Jump to Iodine binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Iodine where determined in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1, PDB code: 4tjv:
Jump to Iodine binding site number: 1; 2; 3; 4; 5; 6;
Iodine binding site 1 out of 6 in 4tjv
Go back to
Iodine binding site 1 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
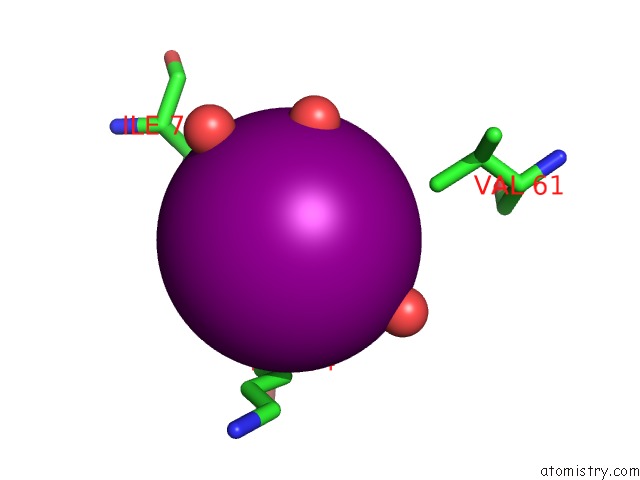
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
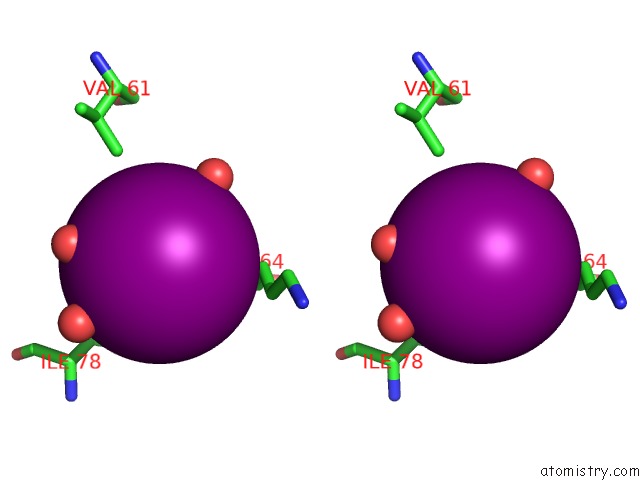
Iodine binding site 2 out of 6 in 4tjv
Go back to
Iodine binding site 2 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
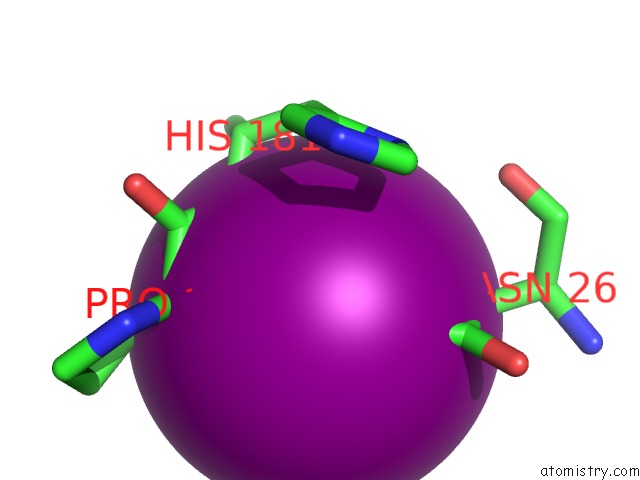
Iodine binding site 3 out of 6 in 4tjv
Go back to
Iodine binding site 3 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
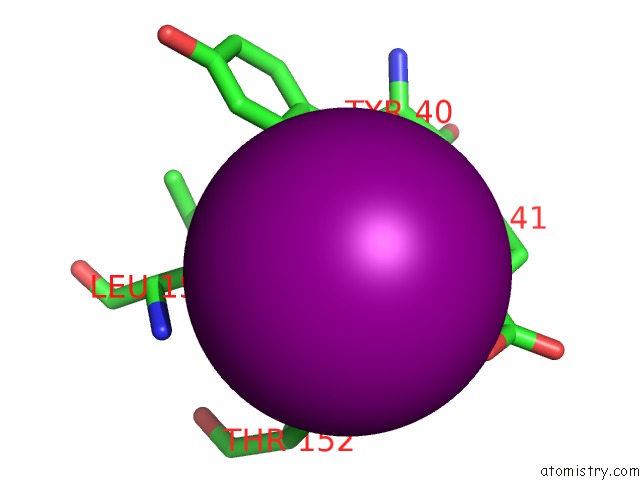
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
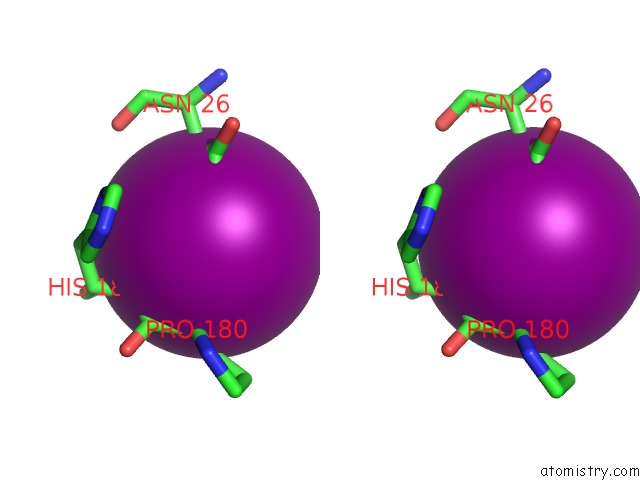
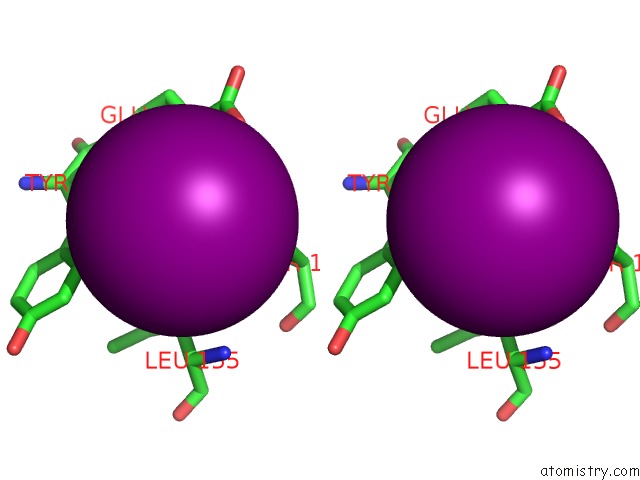
Iodine binding site 4 out of 6 in 4tjv
Go back to
Iodine binding site 4 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
Mono view
Stereo pair view

Mono view
Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
Iodine binding site 5 out of 6 in 4tjv
Go back to
Iodine binding site 5 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
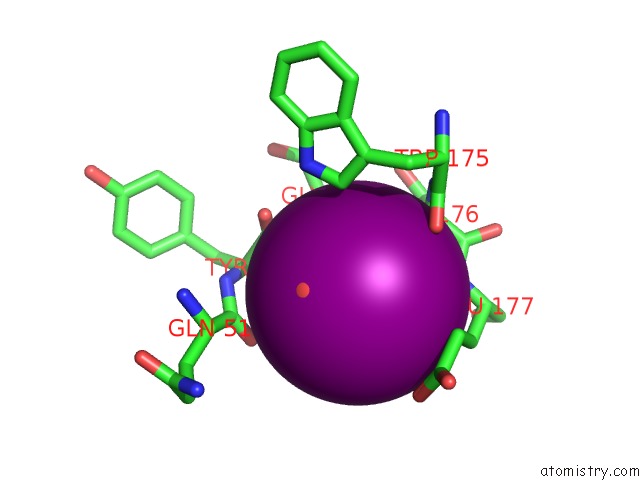
Mono view
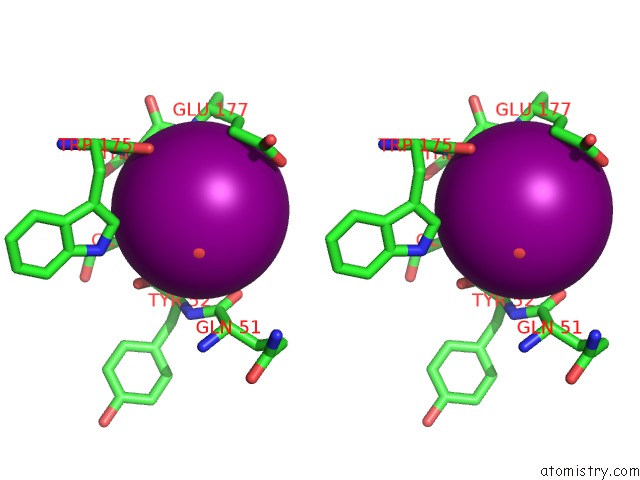
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 5 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
Iodine binding site 6 out of 6 in 4tjv
Go back to
Iodine binding site 6 out
of 6 in the Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 6 of Crystal Structure of Protease-Associated Domain of Arabidopsis Vacuolar Sorting Receptor 1 within 5.0Å range:
|
Reference:
F.Luo,
Y.H.Fong,
Y.Zeng,
J.Shen,
L.Jiang,
K.B.Wong.
How Vacuolar Sorting Receptor Proteins Interact with Their Cargo Proteins: Crystal Structures of Apo and Cargo-Bound Forms of the Protease-Associated Domain From An Arabidopsis Vacuolar Sorting Receptor. Plant Cell V. 26 3693 2014.
ISSN: ESSN 1532-298X
PubMed: 25271241
DOI: 10.1105/TPC.114.129940
Page generated: Sun Aug 11 19:49:41 2024
ISSN: ESSN 1532-298X
PubMed: 25271241
DOI: 10.1105/TPC.114.129940
Last articles
F in 7GP8F in 7GOR
F in 7GOV
F in 7GON
F in 7GOB
F in 7GO1
F in 7GO7
F in 7GN2
F in 7GMZ
F in 7GM4