Iodine »
PDB 5sb0-5w1i »
5tmv »
Iodine in PDB 5tmv: Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate
Protein crystallography data
The structure of Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate, PDB code: 5tmv
was solved by
J.C.Nwachukwu,
R.Erumbi,
S.Srinivasan,
N.E.Bruno,
J.Nowak,
T.Izard,
D.J.Kojetin,
O.Elemento,
J.A.Katzenellenbogen,
K.W.Nettles,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.23 / 2.38 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.390, 81.520, 58.360, 90.00, 110.61, 90.00 |
| R / Rfree (%) | 19.3 / 25.2 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate
(pdb code 5tmv). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 2 binding sites of Iodine where determined in the Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate, PDB code: 5tmv:
Jump to Iodine binding site number: 1; 2;
In total 2 binding sites of Iodine where determined in the Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate, PDB code: 5tmv:
Jump to Iodine binding site number: 1; 2;
Iodine binding site 1 out of 2 in 5tmv
Go back to
Iodine binding site 1 out
of 2 in the Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate
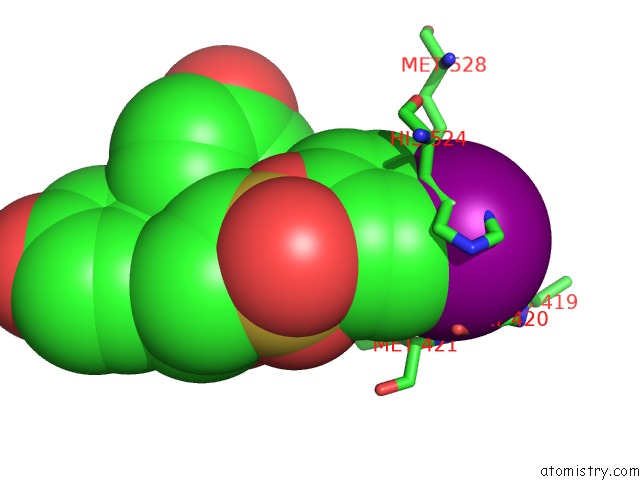
Mono view
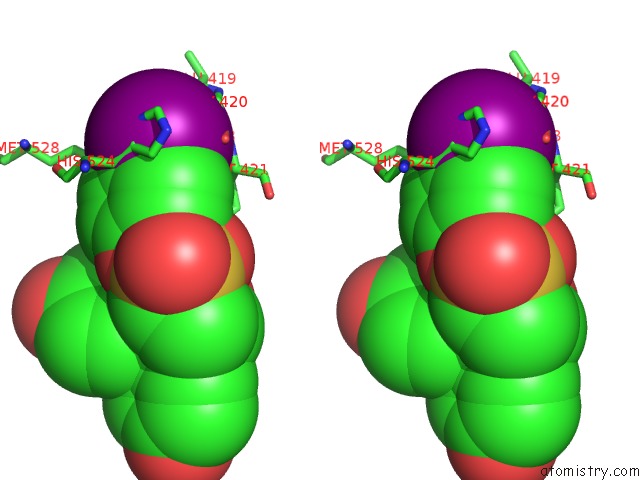
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate within 5.0Å range:
|
Iodine binding site 2 out of 2 in 5tmv
Go back to
Iodine binding site 2 out
of 2 in the Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate
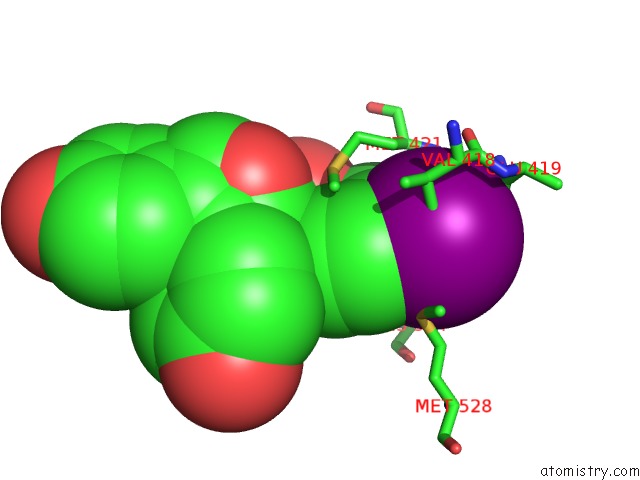
Mono view
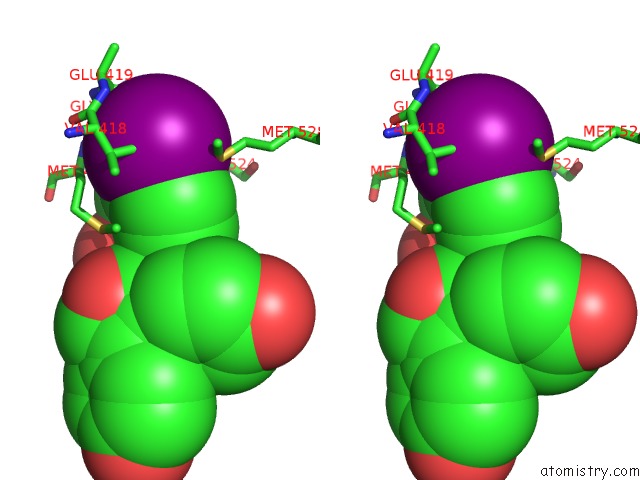
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of the Er-Alpha Ligand-Binding Domain (Y537S) in Complex with the Obhs Analog, 4-Iodophenyl (1S,2R,4S)-5,6-Bis(4- Hydroxyphenyl)-7-Oxabicyclo[2.2.1]Hept-5-Ene-2-Sulfonate within 5.0Å range:
|
Reference:
J.C.Nwachukwu,
S.Srinivasan,
N.E.Bruno,
J.Nowak,
N.J.Wright,
F.Minutolo,
E.S.Rangarajan,
T.Izard,
X.Q.Yao,
B.J.Grant,
D.J.Kojetin,
O.Elemento,
J.A.Katzenellenbogen,
K.W.Nettles.
Systems Structural Biology Analysis of Ligand Effects on Er Alpha Predicts Cellular Response to Environmental Estrogens and Anti-Hormone Therapies. Cell Chem Biol V. 24 35 2017.
ISSN: ESSN 2451-9456
PubMed: 28042045
DOI: 10.1016/J.CHEMBIOL.2016.11.014
Page generated: Sun Aug 11 21:52:56 2024
ISSN: ESSN 2451-9456
PubMed: 28042045
DOI: 10.1016/J.CHEMBIOL.2016.11.014
Last articles
Cl in 3UCNCl in 3UCK
Cl in 3UBT
Cl in 3UC7
Cl in 3UCJ
Cl in 3UBK
Cl in 3UBL
Cl in 3UA5
Cl in 3UB5
Cl in 3UA7