Iodine »
PDB 5w4i-6axx »
5zkz »
Iodine in PDB 5zkz: Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
Enzymatic activity of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
All present enzymatic activity of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose:
3.2.1.8;
3.2.1.8;
Protein crystallography data
The structure of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose, PDB code: 5zkz
was solved by
X.Zhang,
Q.Wan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.08 / 1.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.195, 59.109, 69.709, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.3 / 17.6 |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
(pdb code 5zkz). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 3 binding sites of Iodine where determined in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose, PDB code: 5zkz:
Jump to Iodine binding site number: 1; 2; 3;
In total 3 binding sites of Iodine where determined in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose, PDB code: 5zkz:
Jump to Iodine binding site number: 1; 2; 3;
Iodine binding site 1 out of 3 in 5zkz
Go back to
Iodine binding site 1 out
of 3 in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
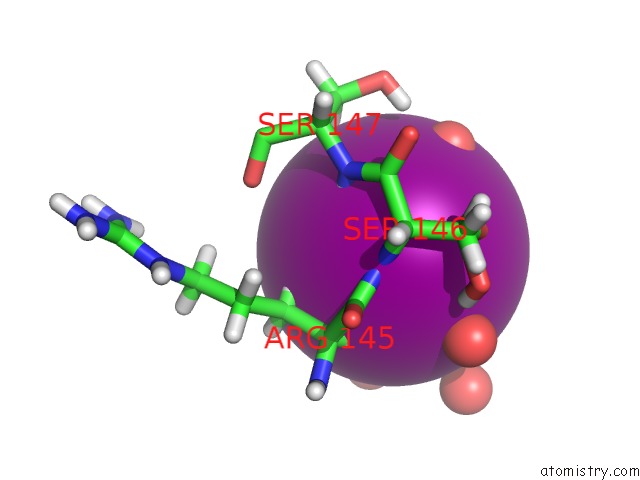
Mono view
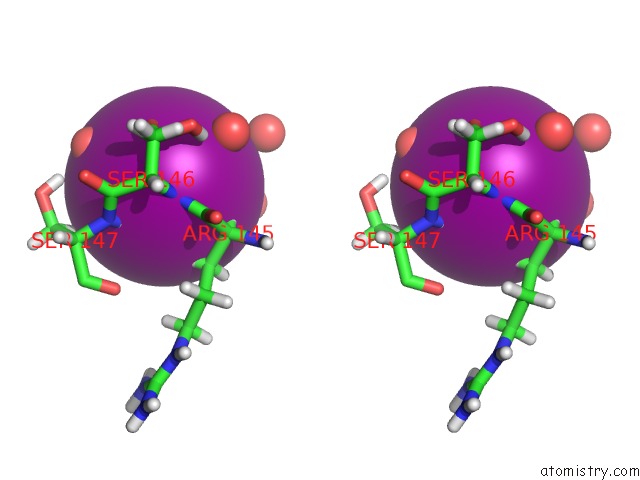
Stereo pair view

Mono view
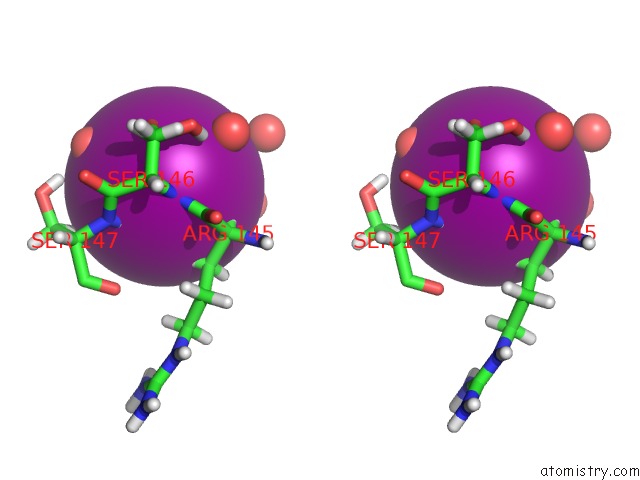
Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose within 5.0Å range:
|
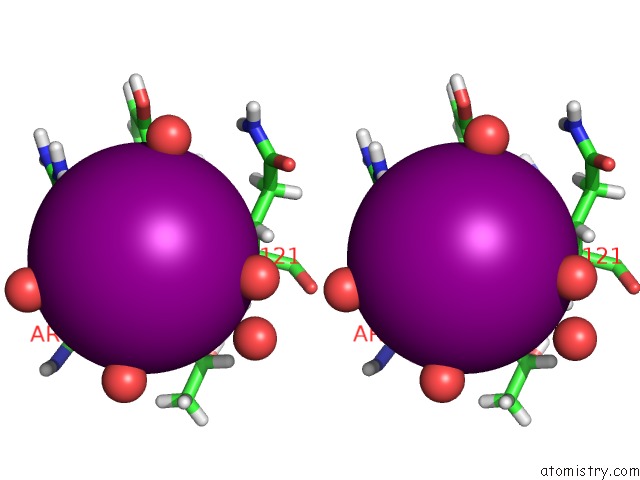
Iodine binding site 2 out of 3 in 5zkz
Go back to
Iodine binding site 2 out
of 3 in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
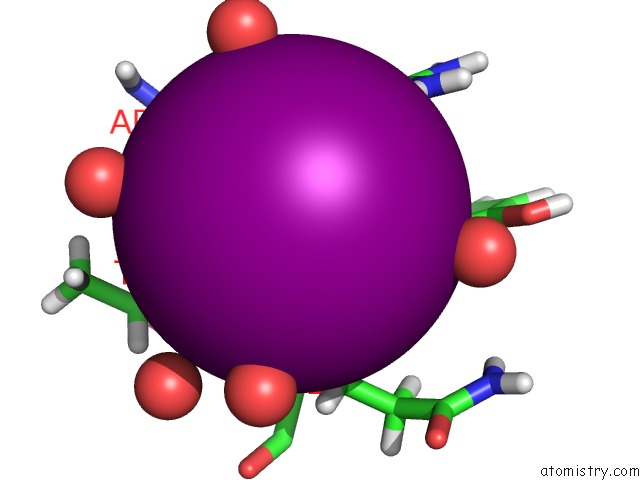
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose within 5.0Å range:
|
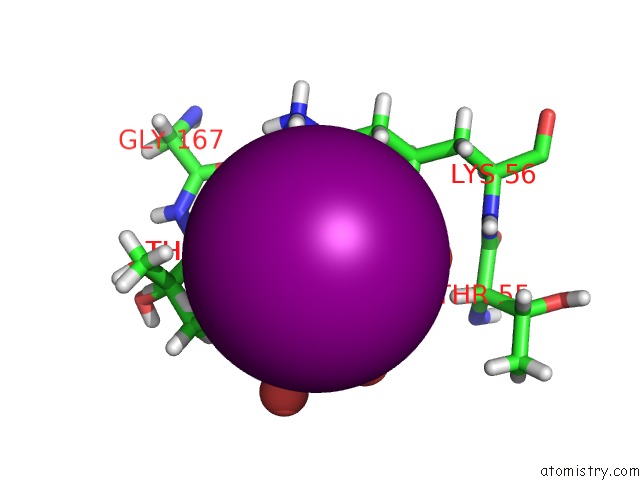
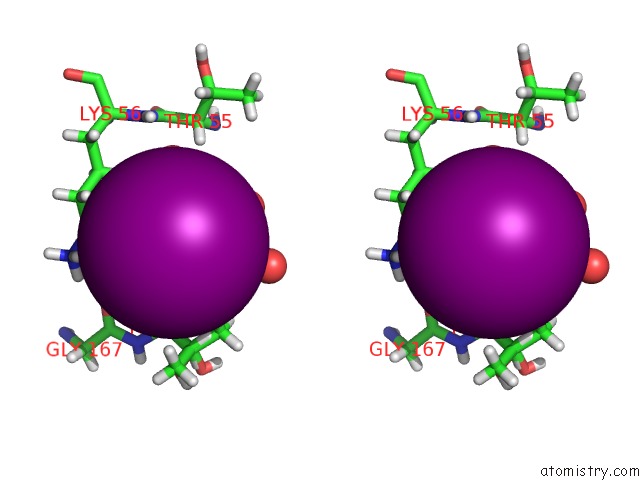
Iodine binding site 3 out of 3 in 5zkz
Go back to
Iodine binding site 3 out
of 3 in the Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structures of Mutant Endo-Beta-1,4-Xylanase II(Y77F) Complexed with Xylotriose within 5.0Å range:
|
Reference:
Q.Wan,
X.Zhang.
Crystal Structures of Endo-Beta-1,4-Xylanase II To Be Published.
Page generated: Sun Aug 11 22:27:04 2024
Last articles
F in 4DBUF in 4DHM
F in 4DEB
F in 4DC3
F in 4D8C
F in 4D83
F in 4DBQ
F in 4DBN
F in 4DAN
F in 4DA4