Iodine »
PDB 6s0t-6vx2 »
6tmb »
Iodine in PDB 6tmb: Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
Protein crystallography data
The structure of Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases, PDB code: 6tmb
was solved by
H.-K.S.Leiros,
T.Christopeit,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 31.60 / 1.50 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 100.590, 79.026, 67.235, 90.00, 130.09, 90.00 |
| R / Rfree (%) | 13.9 / 16.8 |
Other elements in 6tmb:
The structure of Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases also contains other interesting chemical elements:
| Zinc | (Zn) | 6 atoms |
| Chlorine | (Cl) | 4 atoms |
Iodine Binding Sites:
The binding sites of Iodine atom in the Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
(pdb code 6tmb). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total only one binding site of Iodine was determined in the Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases, PDB code: 6tmb:
In total only one binding site of Iodine was determined in the Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases, PDB code: 6tmb:
Iodine binding site 1 out of 1 in 6tmb
Go back to
Iodine binding site 1 out
of 1 in the Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
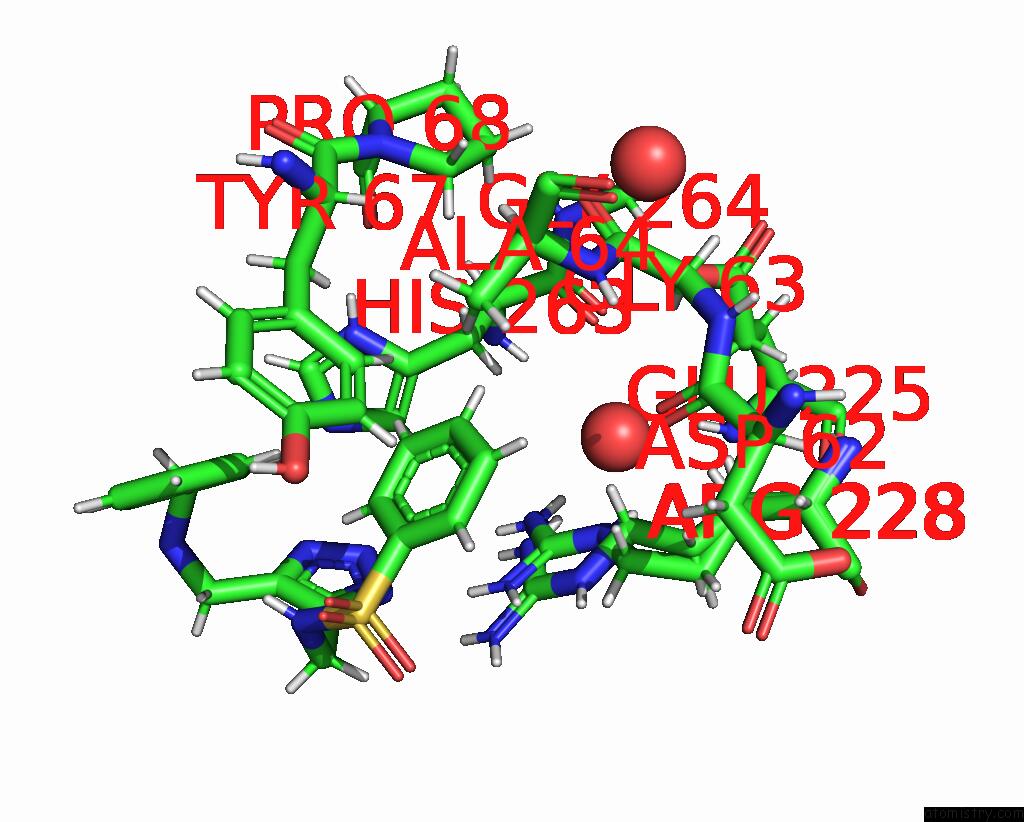
Mono view
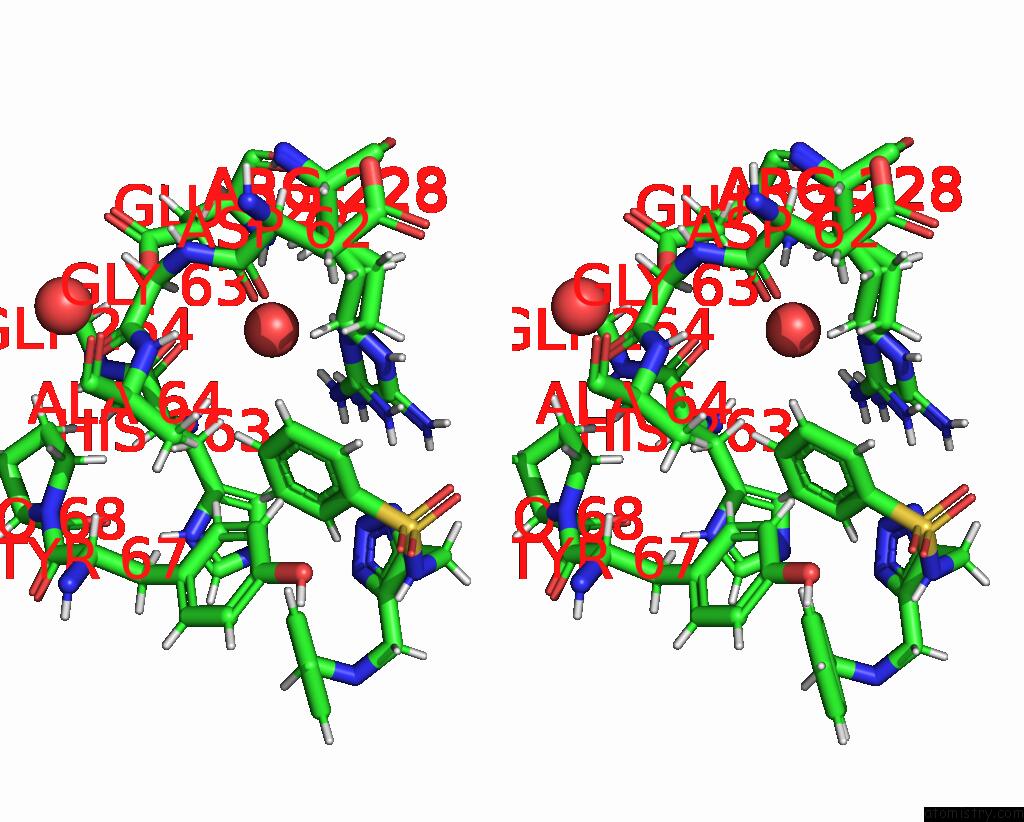
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Vim-2_1DI-Triazole Inhibitors with Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases within 5.0Å range:
|
Reference:
H.-K.S.Leiros,
H.-K.S.Leiros,
T.Christopeit.
N/A N/A.
Page generated: Fri Aug 8 22:18:51 2025
Last articles
K in 1JFVK in 1JF8
K in 1JDR
K in 1JCI
K in 1JBS
K in 1JBT
K in 1J95
K in 1JBR
K in 1J5Y
K in 1IS8