Iodine »
PDB 7lxr-7px1 »
7o2p »
Iodine in PDB 7o2p: Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
Protein crystallography data
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756, PDB code: 7o2p
was solved by
K.Zrubek,
G.Sandrone,
C.D.Cukier,
A.Stevenazzi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.483, 90.753, 71.964, 90, 95.64, 90 |
| R / Rfree (%) | 19.5 / 26.1 |
Other elements in 7o2p:
The structure of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756 also contains other interesting chemical elements:
| Potassium | (K) | 4 atoms |
| Zinc | (Zn) | 2 atoms |
Iodine Binding Sites:
The binding sites of Iodine atom in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
(pdb code 7o2p). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 4 binding sites of Iodine where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756, PDB code: 7o2p:
Jump to Iodine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iodine where determined in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756, PDB code: 7o2p:
Jump to Iodine binding site number: 1; 2; 3; 4;
Iodine binding site 1 out of 4 in 7o2p
Go back to
Iodine binding site 1 out
of 4 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
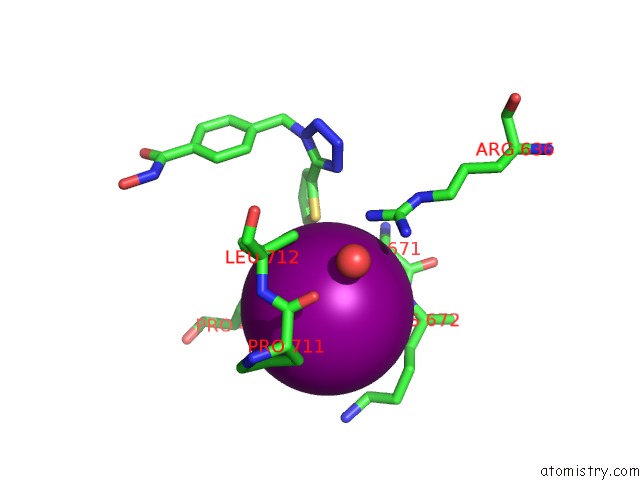
Mono view
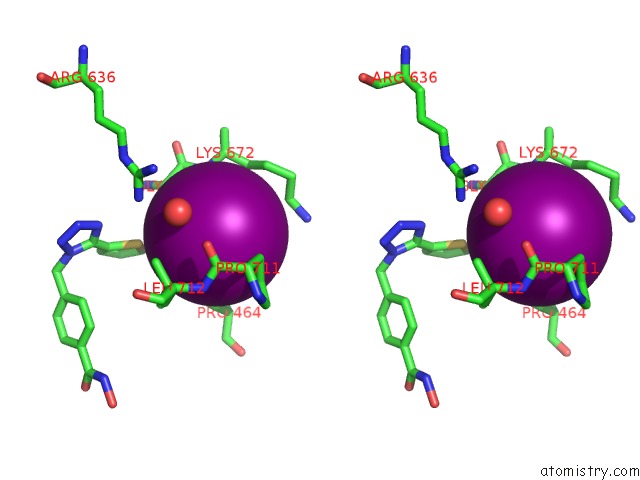
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756 within 5.0Å range:
|
Iodine binding site 2 out of 4 in 7o2p
Go back to
Iodine binding site 2 out
of 4 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
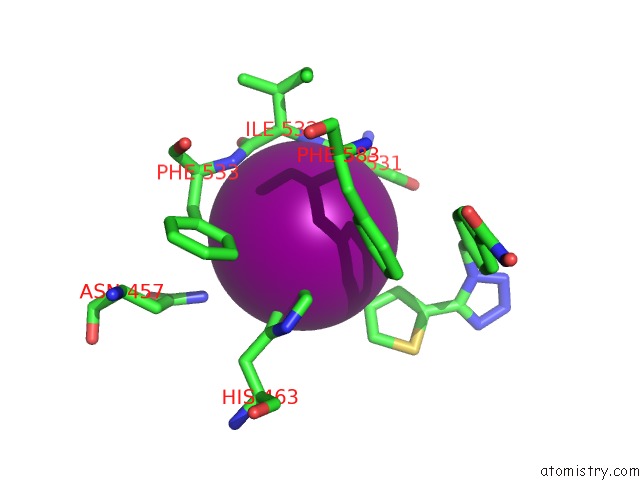
Mono view
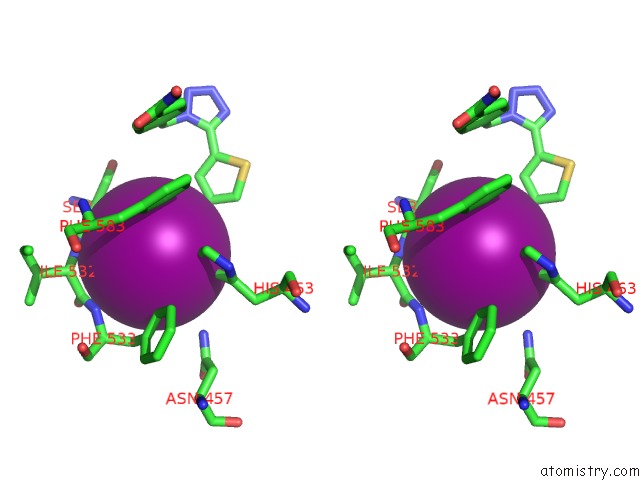
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756 within 5.0Å range:
|
Iodine binding site 3 out of 4 in 7o2p
Go back to
Iodine binding site 3 out
of 4 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
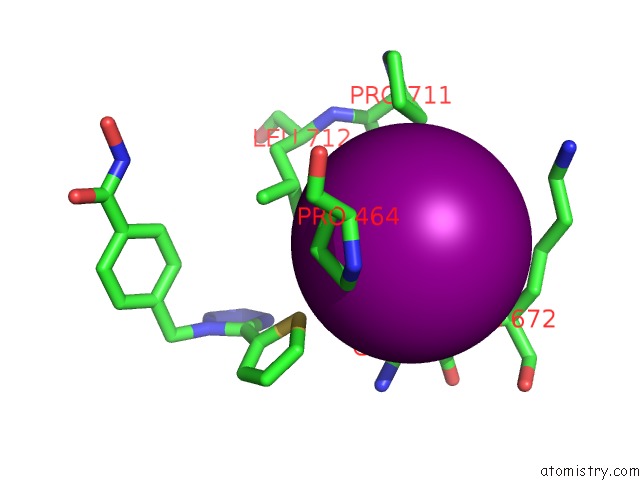
Mono view
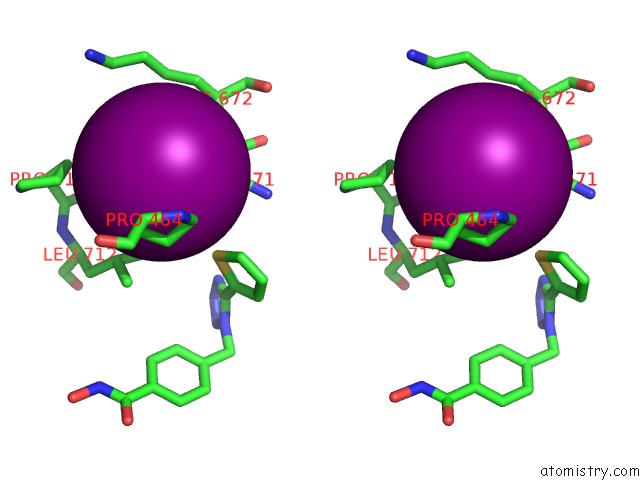
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756 within 5.0Å range:
|
Iodine binding site 4 out of 4 in 7o2p
Go back to
Iodine binding site 4 out
of 4 in the Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756
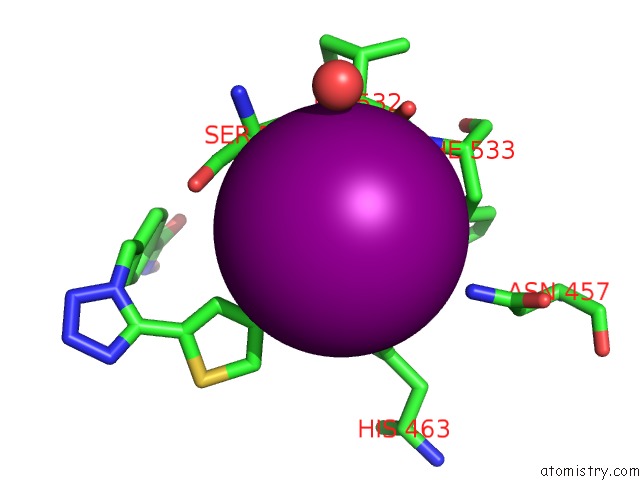
Mono view
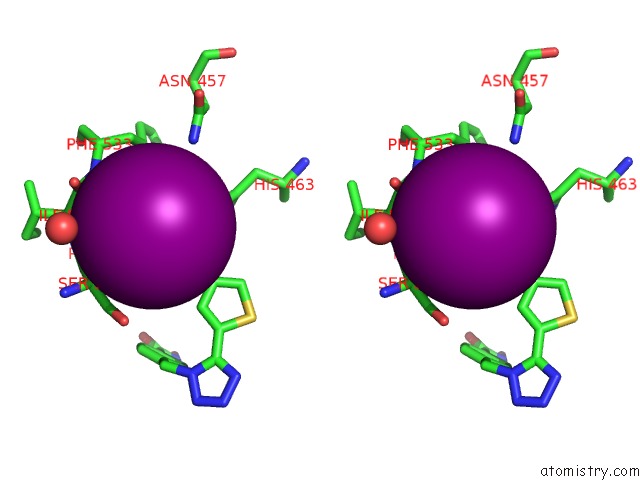
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Crystal Structure of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 in Complex with ITF3756 within 5.0Å range:
|
Reference:
G.Sandrone,
C.D.Cukier,
K.Zrubek,
M.Marchini,
B.Vergani,
G.Caprini,
G.Fossati,
C.Steinkuhler,
A.Stevenazzi.
Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors Acs Med.Chem.Lett. 2021.
ISSN: ISSN 1948-5875
DOI: 10.1021/ACSMEDCHEMLETT.1C00425
Page generated: Fri Aug 8 23:29:43 2025
ISSN: ISSN 1948-5875
DOI: 10.1021/ACSMEDCHEMLETT.1C00425
Last articles
K in 2GHKK in 2GHS
K in 2GDI
K in 2GHH
K in 2G6P
K in 2G4V
K in 2G4N
K in 2FZW
K in 2FZE
K in 2FXI