Iodine »
PDB 7px1-7se4 »
7r4f »
Iodine in PDB 7r4f: Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map)
Enzymatic activity of Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map)
All present enzymatic activity of Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map):
1.6.5.3; 1.6.99.3; 7.1.1.2;
1.6.5.3; 1.6.99.3; 7.1.1.2;
Other elements in 7r4f:
The structure of Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map) also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
| Potassium | (K) | 1 atom |
| Iron | (Fe) | 28 atoms |
| Chlorine | (Cl) | 1 atom |
| Zinc | (Zn) | 1 atom |
Iodine Binding Sites:
The binding sites of Iodine atom in the Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map)
(pdb code 7r4f). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total only one binding site of Iodine was determined in the Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map), PDB code: 7r4f:
In total only one binding site of Iodine was determined in the Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map), PDB code: 7r4f:
Iodine binding site 1 out of 1 in 7r4f
Go back to
Iodine binding site 1 out
of 1 in the Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map)
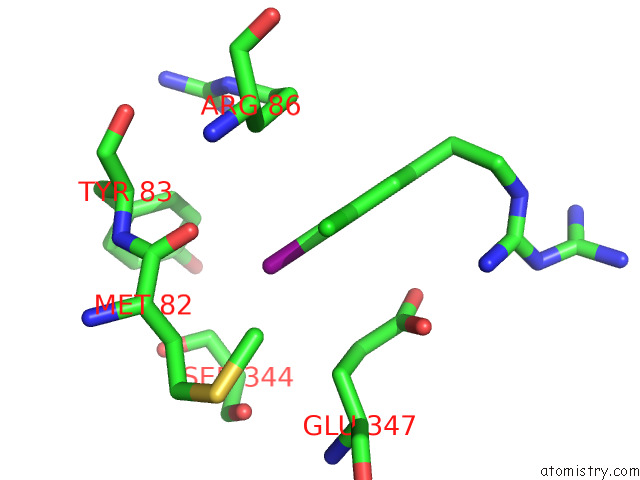
Mono view
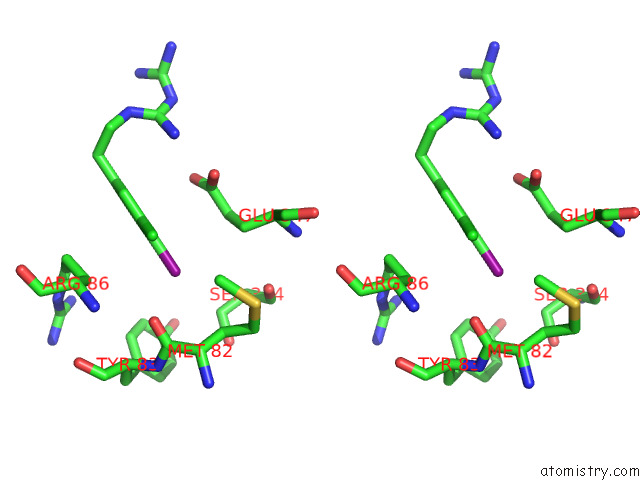
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Bovine Complex I in the Presence of IM1761092, Slack Class I (Composite Map) within 5.0Å range:
|
Reference:
H.R.Bridges,
J.N.Blaza,
Z.Yin,
I.Chung,
M.N.Pollak,
J.Hirst.
Structural Basis of Mammalian Respiratory Complex I Inhibition By Medicinal Biguanides. Science V. 379 351 2023.
ISSN: ESSN 1095-9203
PubMed: 36701435
DOI: 10.1126/SCIENCE.ADE3332
Page generated: Mon Aug 12 02:03:43 2024
ISSN: ESSN 1095-9203
PubMed: 36701435
DOI: 10.1126/SCIENCE.ADE3332
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1