Iodine »
PDB 8a24-8e5b »
8cnz »
Iodine in PDB 8cnz: Mmlare-[4FE-4S] Phased By Fe-Sad
Protein crystallography data
The structure of Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz
was solved by
L.Pecqueur,
P.Zecchin,
B.Golinelli-Pimpaneau,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 55.37 / 3.60 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 187.516, 209.215, 195.799, 90, 90, 90 |
| R / Rfree (%) | 25.3 / 28 |
Other elements in 8cnz:
The structure of Mmlare-[4FE-4S] Phased By Fe-Sad also contains other interesting chemical elements:
| Iron | (Fe) | 28 atoms |
| Chlorine | (Cl) | 5 atoms |
Iodine Binding Sites:
The binding sites of Iodine atom in the Mmlare-[4FE-4S] Phased By Fe-Sad
(pdb code 8cnz). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 4 binding sites of Iodine where determined in the Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz:
Jump to Iodine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Iodine where determined in the Mmlare-[4FE-4S] Phased By Fe-Sad, PDB code: 8cnz:
Jump to Iodine binding site number: 1; 2; 3; 4;
Iodine binding site 1 out of 4 in 8cnz
Go back to
Iodine binding site 1 out
of 4 in the Mmlare-[4FE-4S] Phased By Fe-Sad
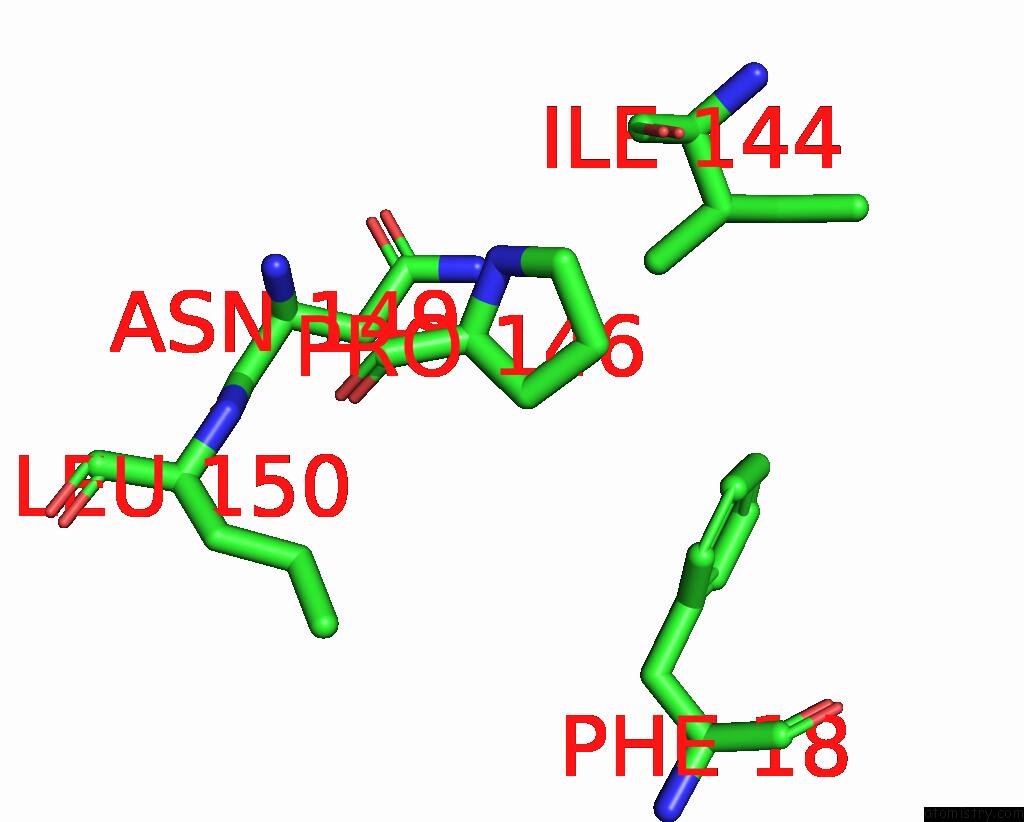
Mono view
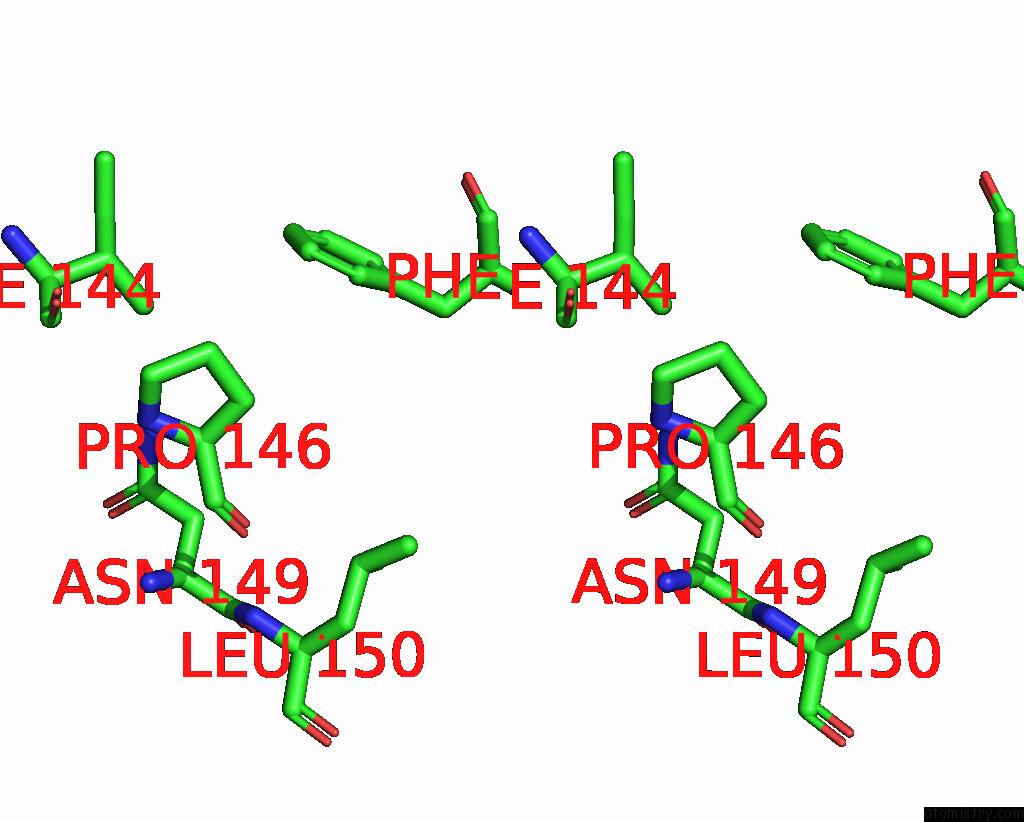
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
Iodine binding site 2 out of 4 in 8cnz
Go back to
Iodine binding site 2 out
of 4 in the Mmlare-[4FE-4S] Phased By Fe-Sad
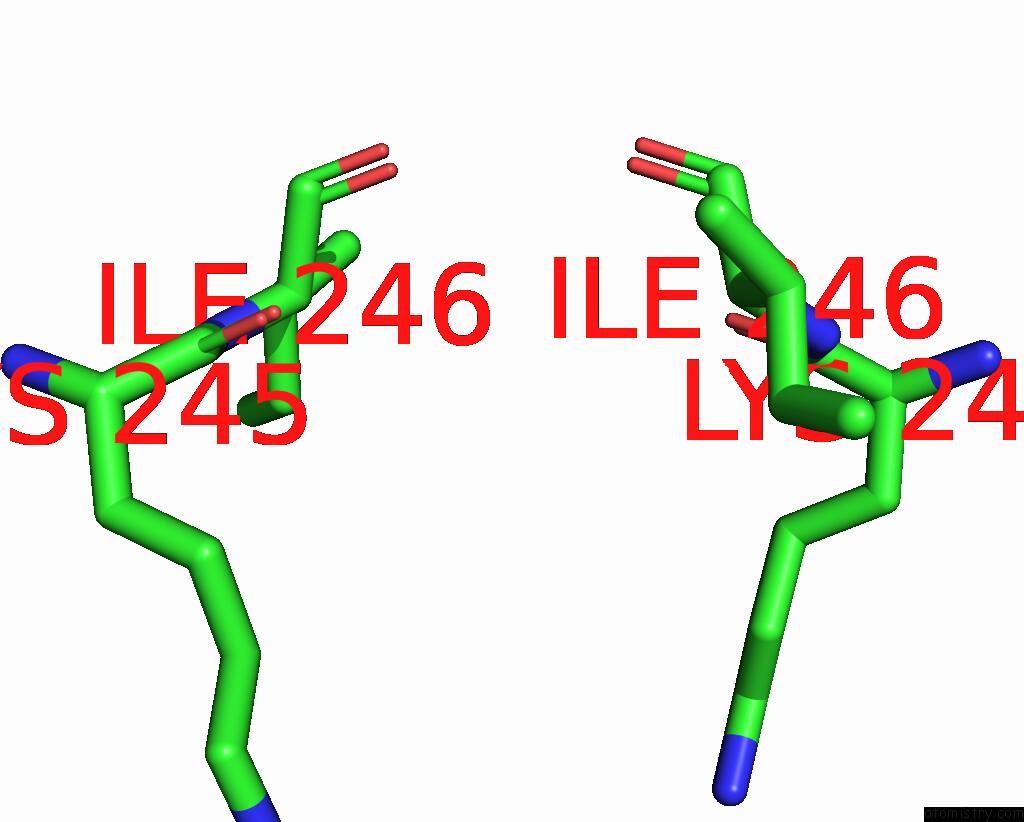
Mono view
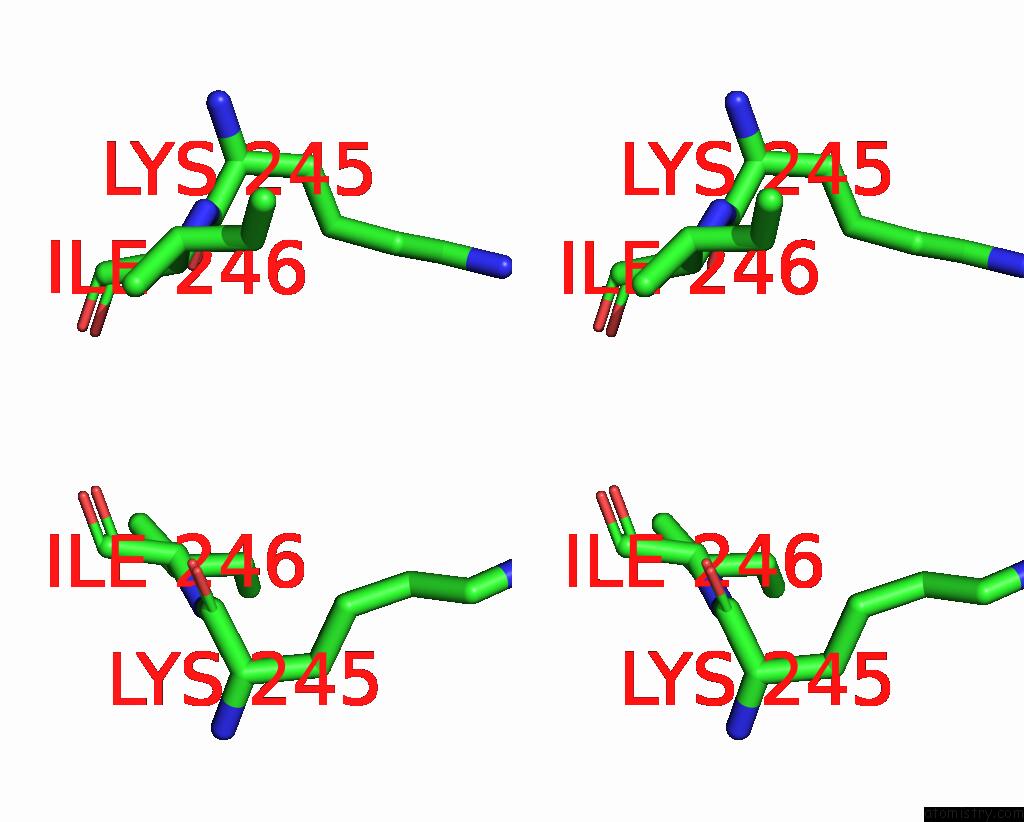
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
Iodine binding site 3 out of 4 in 8cnz
Go back to
Iodine binding site 3 out
of 4 in the Mmlare-[4FE-4S] Phased By Fe-Sad
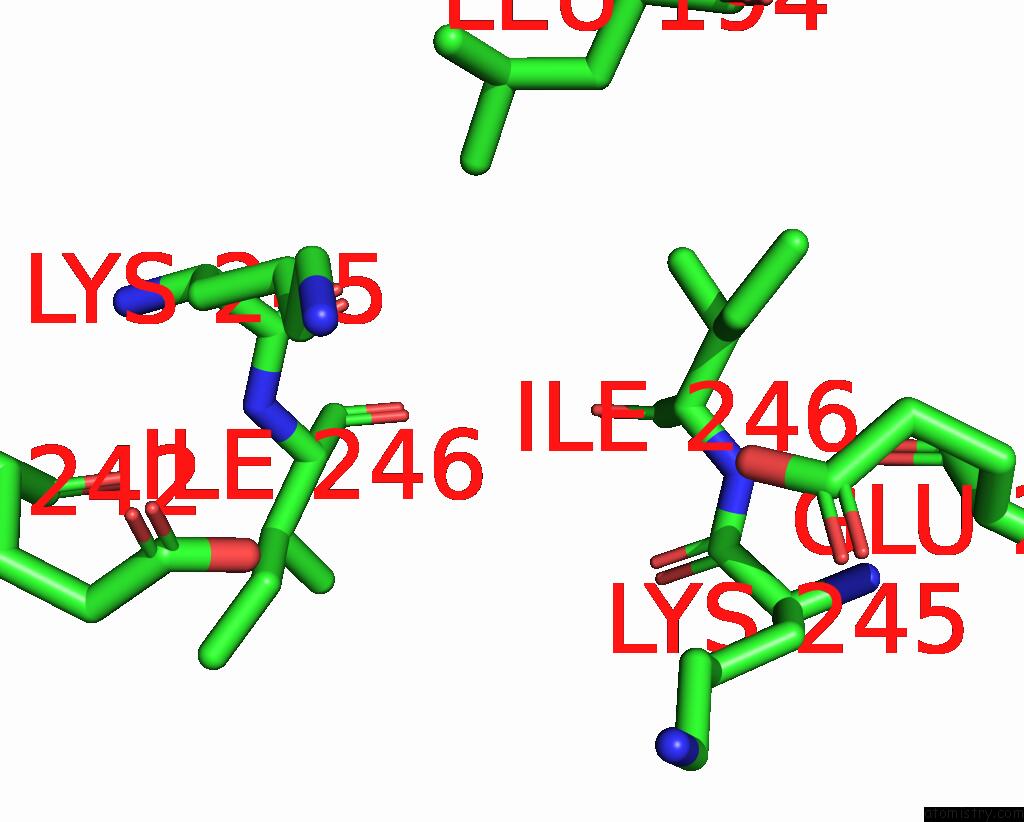
Mono view
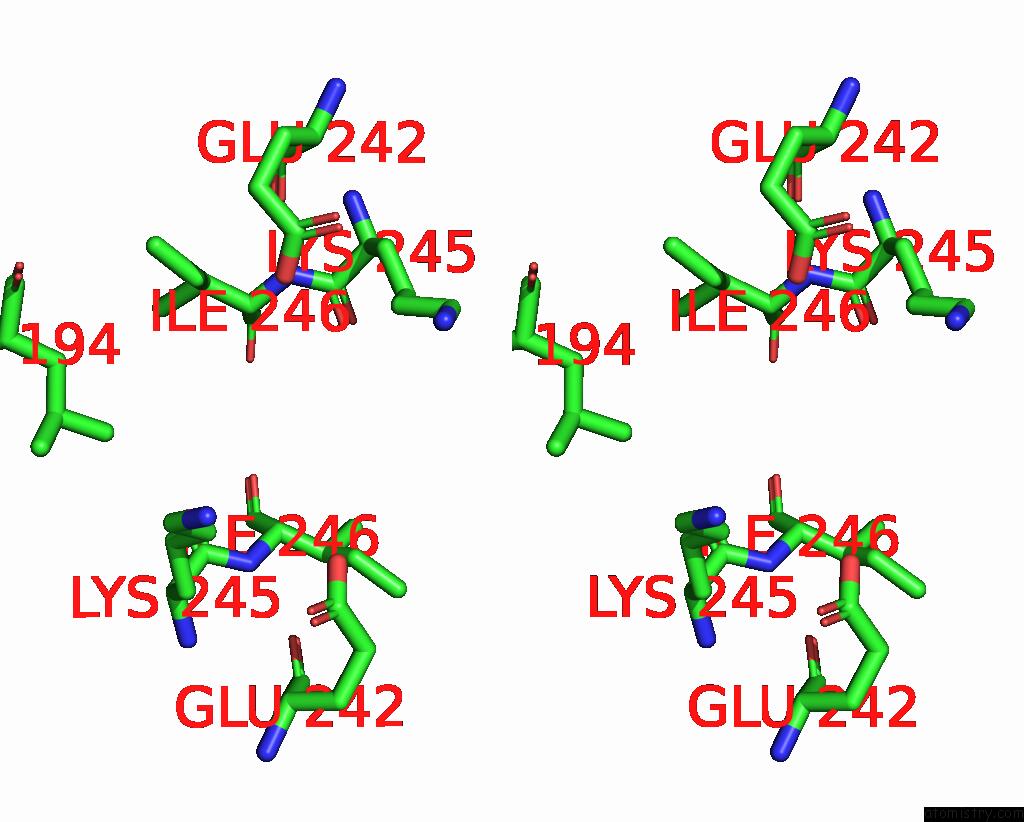
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
Iodine binding site 4 out of 4 in 8cnz
Go back to
Iodine binding site 4 out
of 4 in the Mmlare-[4FE-4S] Phased By Fe-Sad
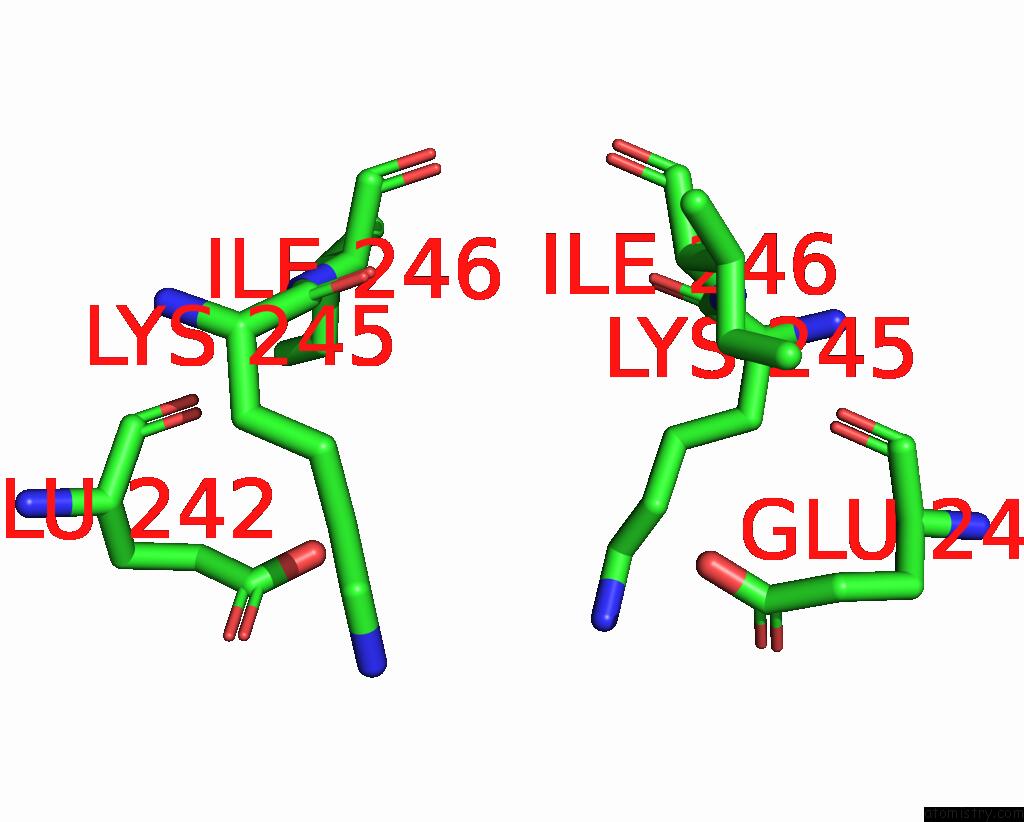
Mono view
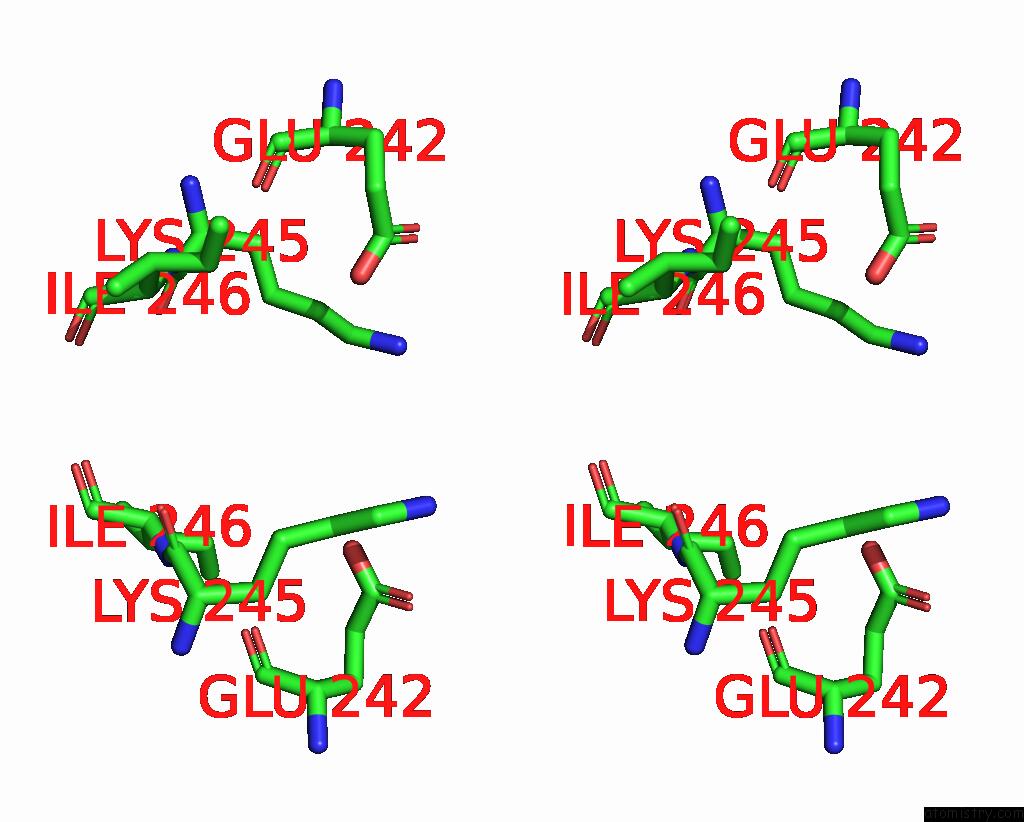
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Mmlare-[4FE-4S] Phased By Fe-Sad within 5.0Å range:
|
Reference:
P.Zecchin,
L.Pecqueur,
J.Oltmanns,
C.Velours,
V.Schunemann,
M.Fontecave,
B.Golinelli-Pimpaneau.
Structure-Based Insights Into the Mechanism of [4FE-4S]-Dependent Sulfur Insertase Lare. Protein Sci. E4874 2023.
ISSN: ESSN 1469-896X
PubMed: 38100250
DOI: 10.1002/PRO.4874
Page generated: Mon Aug 12 02:34:31 2024
ISSN: ESSN 1469-896X
PubMed: 38100250
DOI: 10.1002/PRO.4874
Last articles
Cl in 7TRVCl in 7TSI
Cl in 7TSG
Cl in 7TSH
Cl in 7TRU
Cl in 7TPM
Cl in 7TRT
Cl in 7TRP
Cl in 7TQM
Cl in 7TP6