Iodine »
PDB 4hkl-4jxj »
4izg »
Iodine in PDB 4izg: Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Protein crystallography data
The structure of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product), PDB code: 4izg
was solved by
M.W.Vetting,
R.Toro,
R.Bhosle,
S.R.Wasserman,
L.L.Morisco,
S.Sojitra,
S.Chamala,
A.Kar,
J.Lafleur,
G.Villigas,
B.Evans,
J.Hammonds,
A.Gizzi,
M.Stead,
B.Hillerich,
J.Love,
R.D.Seidel,
J.B.Bonanno,
J.A.Gerlt,
S.C.Almo,
New York Structural Genomics Research Consortium (Nysgrc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 80.70 / 1.70 |
| Space group | P 4 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 117.463, 117.463, 111.049, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.5 / 19.7 |
Other elements in 4izg:
The structure of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) also contains other interesting chemical elements:
| Magnesium | (Mg) | 5 atoms |
Iodine Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20; Page 3, Binding sites: 21 - 28;Binding sites:
The binding sites of Iodine atom in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) (pdb code 4izg). This binding sites where shown within 5.0 Angstroms radius around Iodine atom.In total 28 binding sites of Iodine where determined in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product), PDB code: 4izg:
Jump to Iodine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Iodine binding site 1 out of 28 in 4izg
Go back to
Iodine binding site 1 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
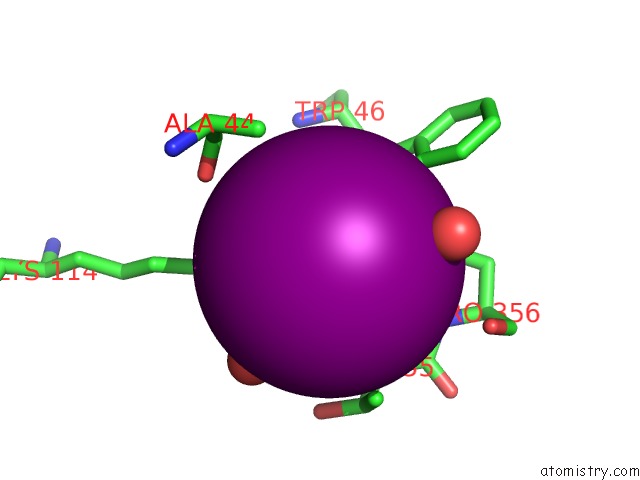
Mono view
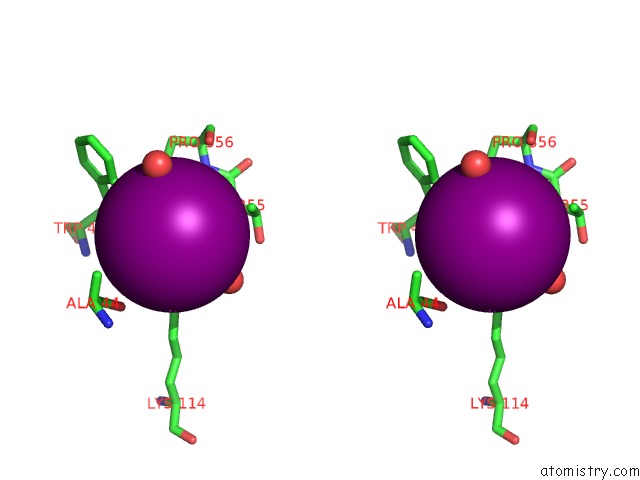
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 2 out of 28 in 4izg
Go back to
Iodine binding site 2 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
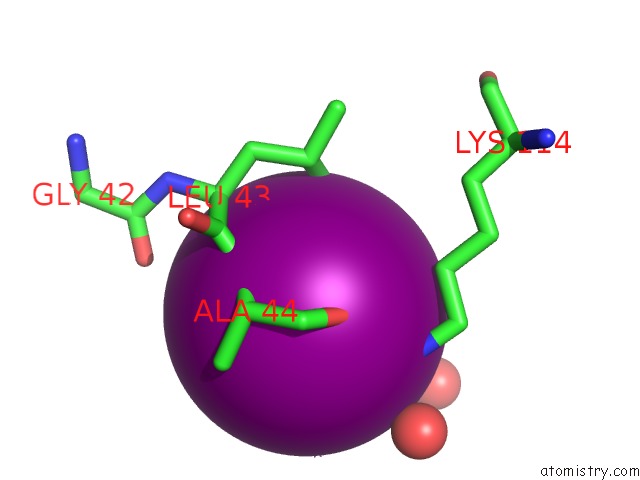
Mono view
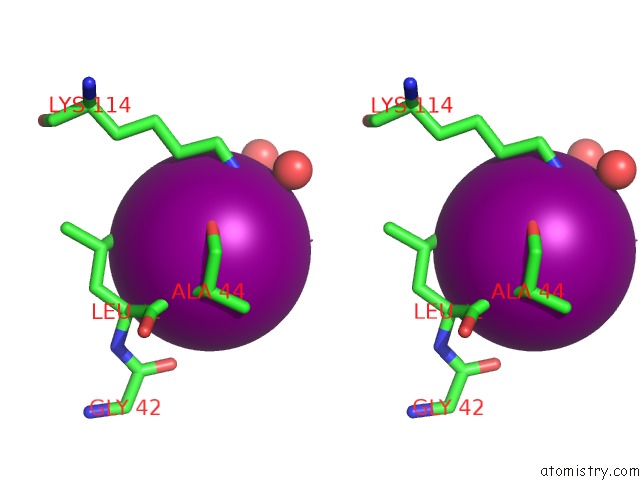
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 3 out of 28 in 4izg
Go back to
Iodine binding site 3 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
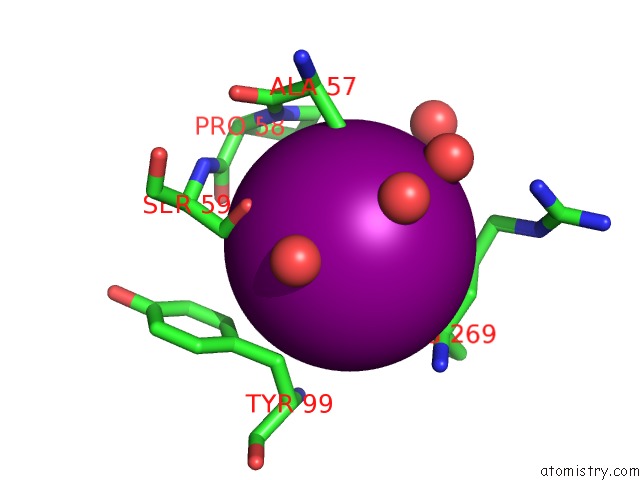
Mono view
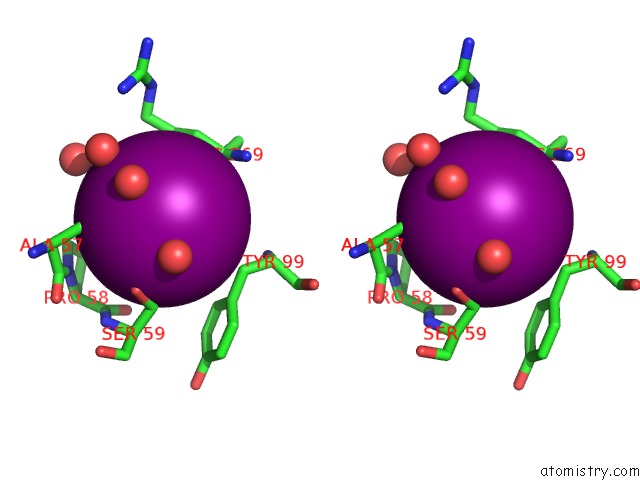
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 4 out of 28 in 4izg
Go back to
Iodine binding site 4 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
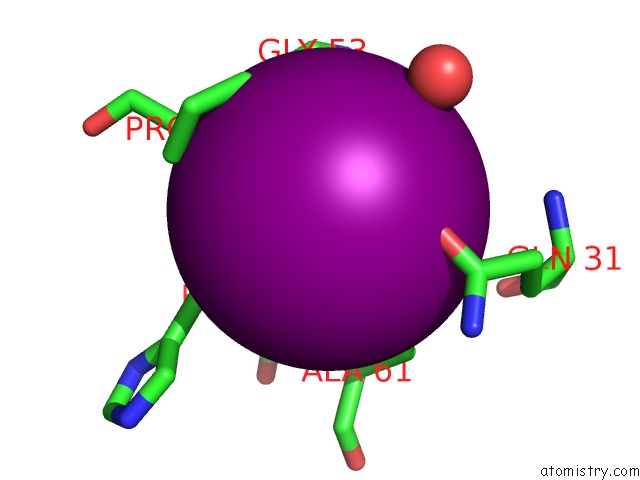
Mono view
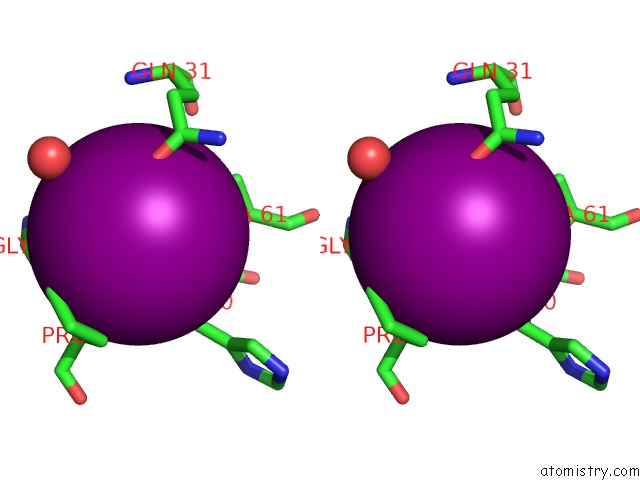
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 5 out of 28 in 4izg
Go back to
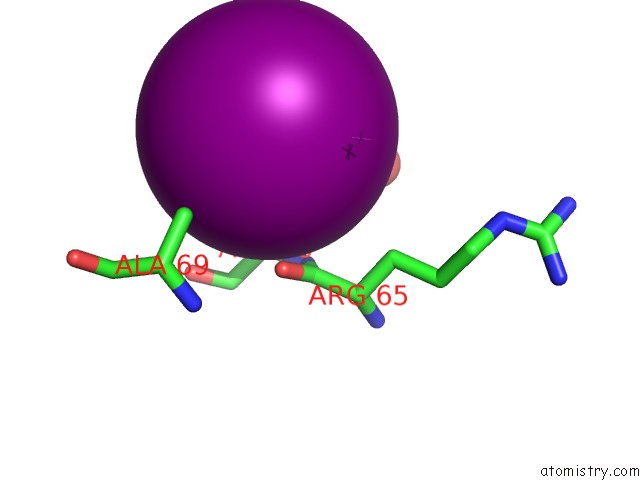
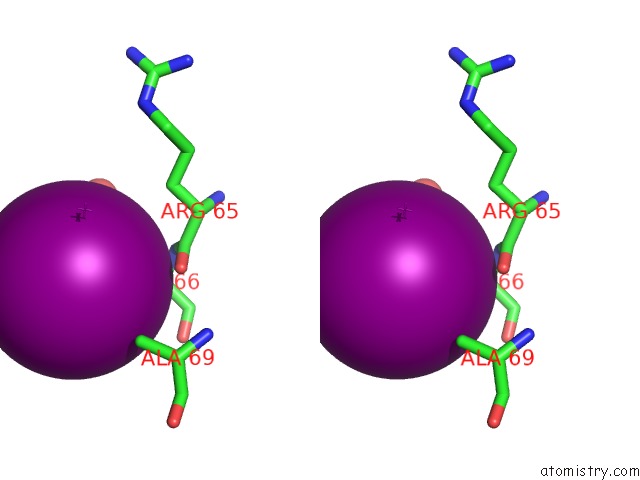
Iodine binding site 5 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 5 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 6 out of 28 in 4izg
Go back to
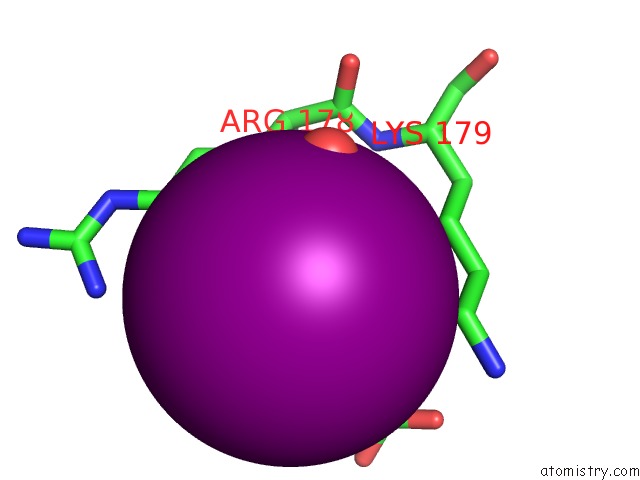
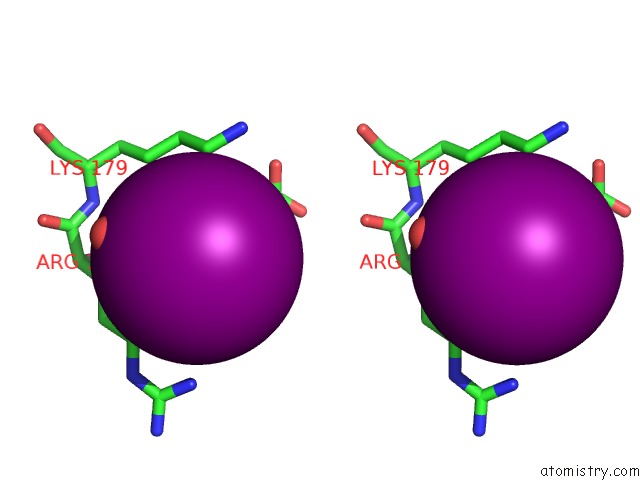
Iodine binding site 6 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 6 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 7 out of 28 in 4izg
Go back to
Iodine binding site 7 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 7 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Iodine binding site 8 out of 28 in 4izg
Go back to
Iodine binding site 8 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 8 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
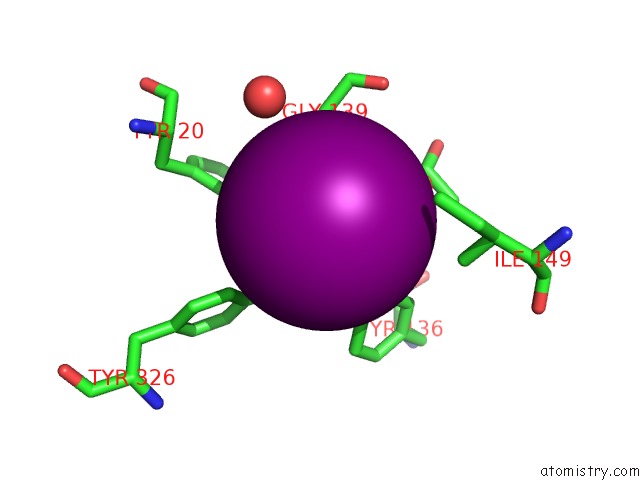
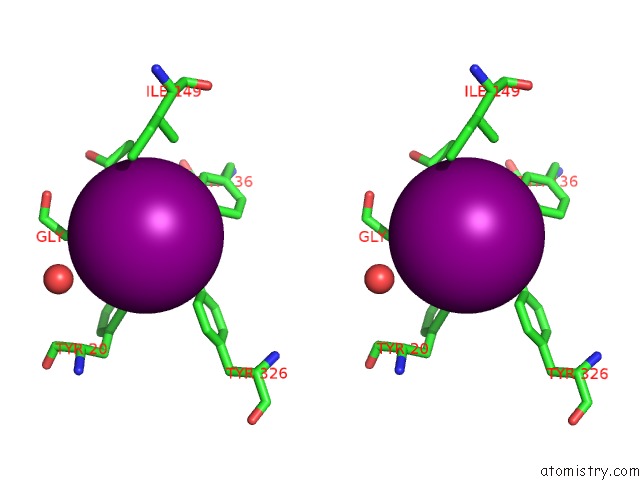
Iodine binding site 9 out of 28 in 4izg
Go back to
Iodine binding site 9 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 9 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
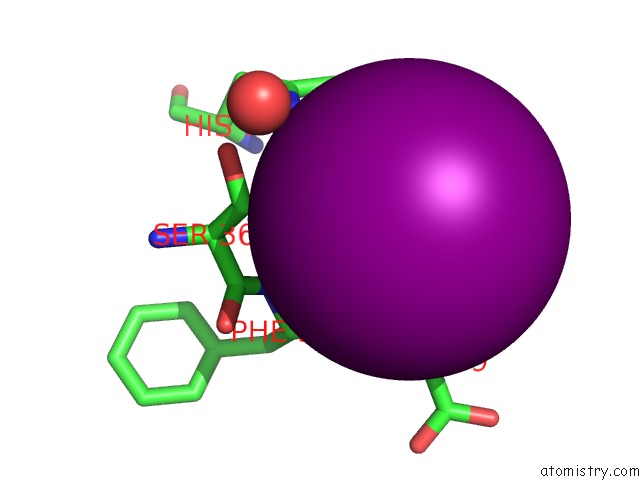
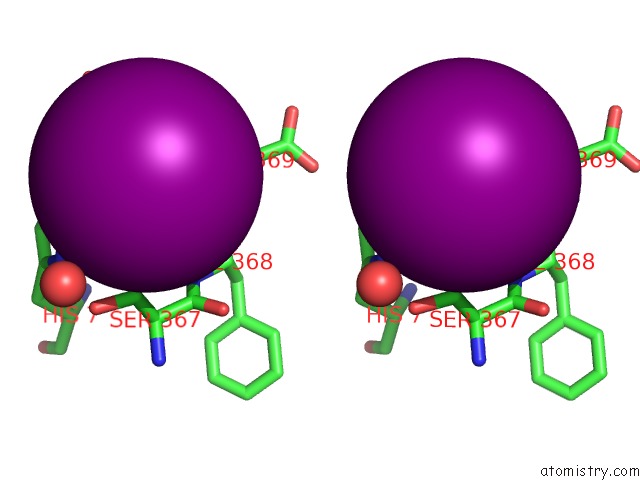
Iodine binding site 10 out of 28 in 4izg
Go back to
Iodine binding site 10 out
of 28 in the Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 10 of Crystal Structure of An Enolase (Mandelate Racemase Subgroup) From Paracococus Denitrificans PD1222 (Target Nysgrc-012907) with Bound Cis-4OH-D-Proline Betaine (Product) within 5.0Å range:
|
Reference:
R.Kumar,
S.Zhao,
M.W.Vetting,
B.M.Wood,
A.Sakai,
K.Cho,
J.Solbiati,
S.C.Almo,
J.V.Sweedler,
M.P.Jacobson,
J.A.Gerlt,
J.E.Cronan.
Prediction and Biochemical Demonstration of A Catabolic Pathway For the Osmoprotectant Proline Betaine. Mbio V. 5 00933 2014.
ISSN: ESSN 2150-7511
PubMed: 24520058
DOI: 10.1128/MBIO.00933-13
Page generated: Fri Aug 8 17:26:50 2025
ISSN: ESSN 2150-7511
PubMed: 24520058
DOI: 10.1128/MBIO.00933-13
Last articles
I in 5F1WI in 5EOJ
I in 5EUZ
I in 5ENM
I in 5ENK
I in 5EEK
I in 5E9O
I in 5EMS
I in 5EIJ
I in 5EI1