Iodine »
PDB 7px1-7se4 »
7qur »
Iodine in PDB 7qur: Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
Iodine Binding Sites:
The binding sites of Iodine atom in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
(pdb code 7qur). This binding sites where shown within
5.0 Angstroms radius around Iodine atom.
In total 9 binding sites of Iodine where determined in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry, PDB code: 7qur:
Jump to Iodine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Iodine where determined in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry, PDB code: 7qur:
Jump to Iodine binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
Iodine binding site 1 out of 9 in 7qur
Go back to
Iodine binding site 1 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 1 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
Iodine binding site 2 out of 9 in 7qur
Go back to
Iodine binding site 2 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
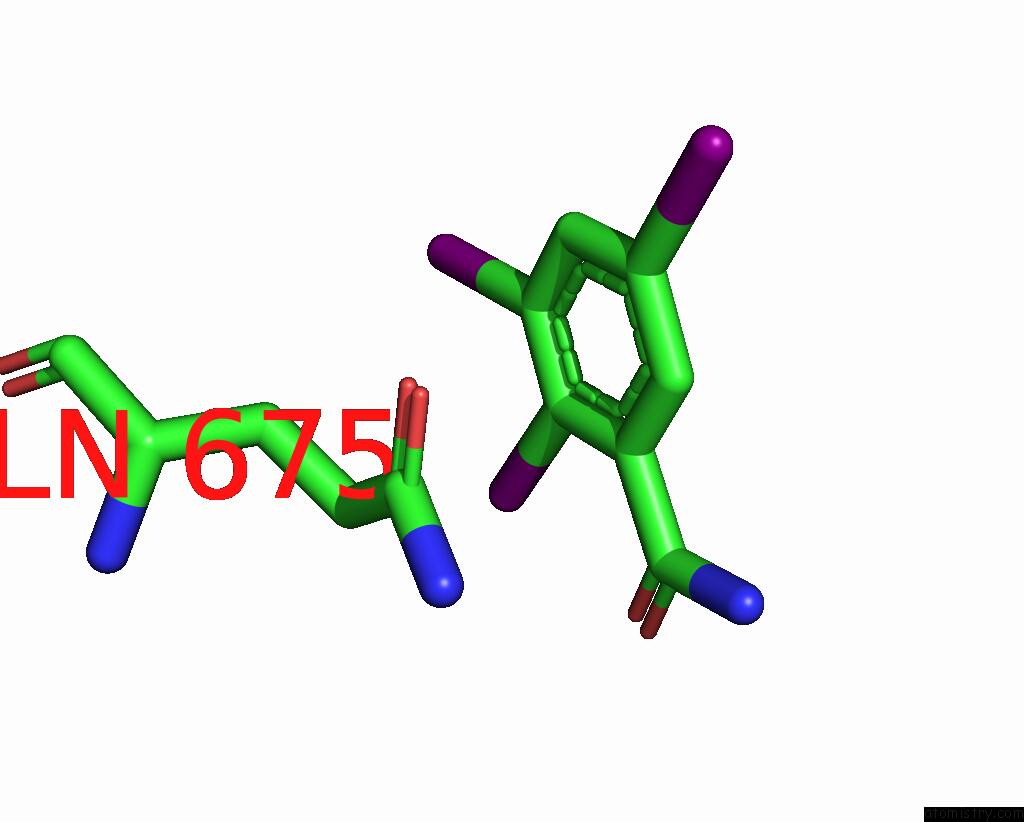
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 2 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
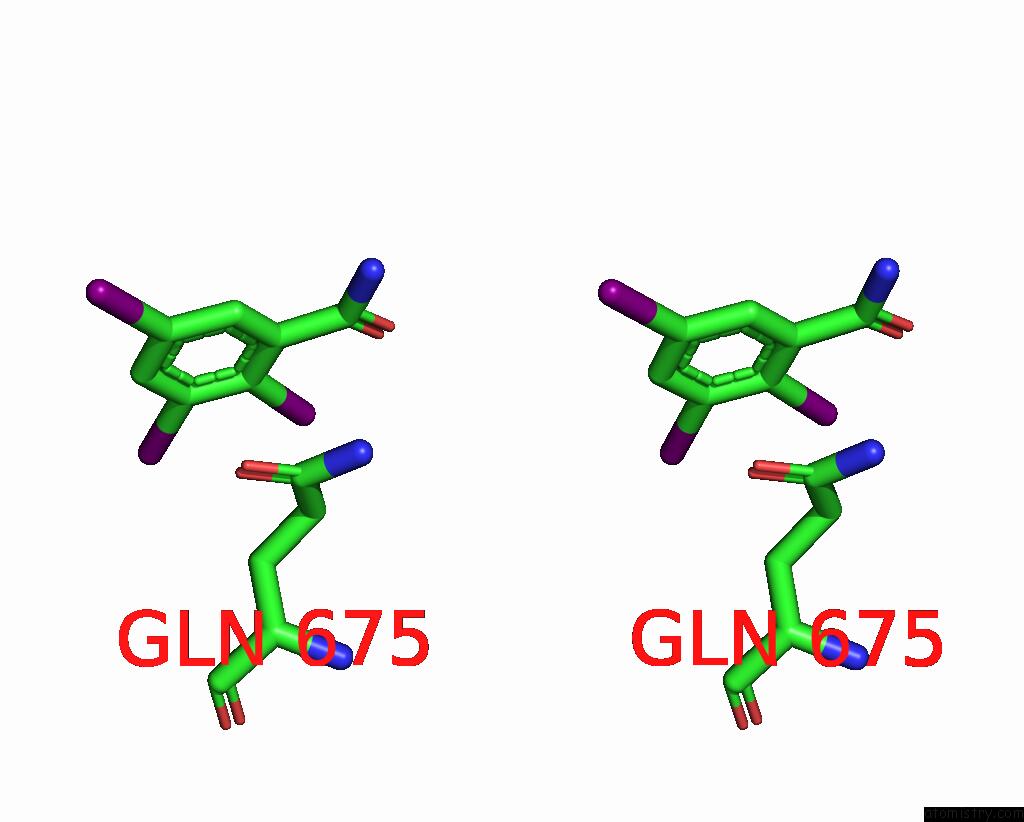
Iodine binding site 3 out of 9 in 7qur
Go back to
Iodine binding site 3 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 3 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
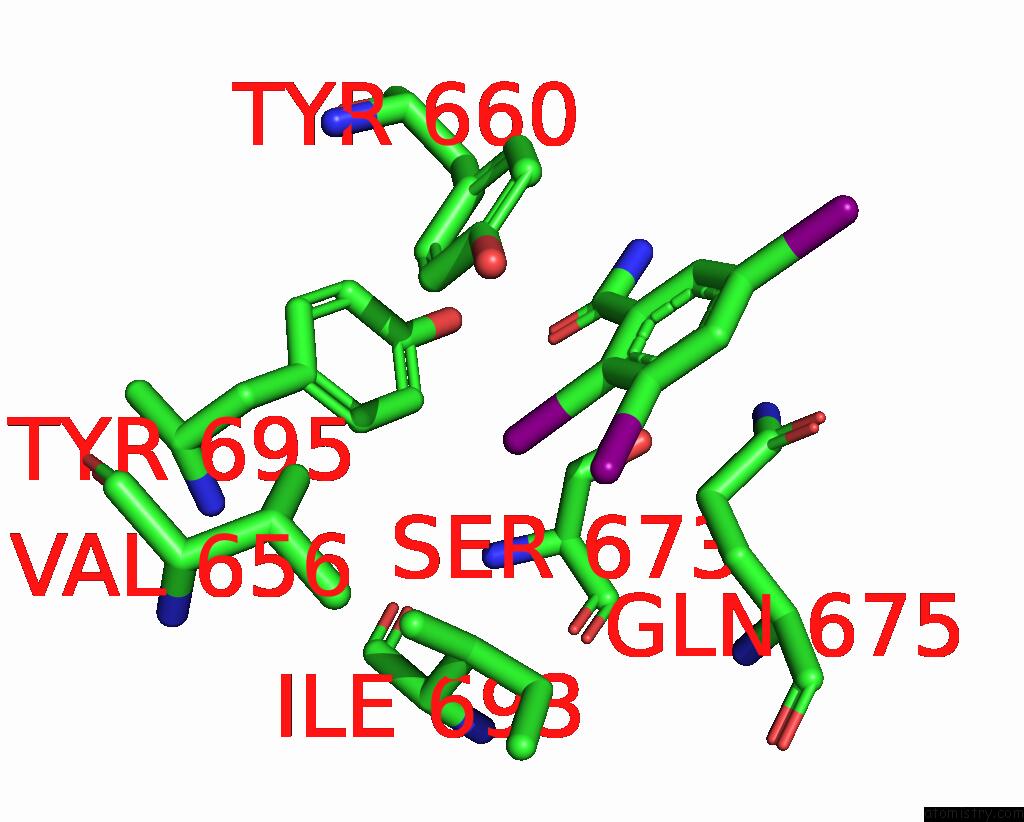
Iodine binding site 4 out of 9 in 7qur
Go back to
Iodine binding site 4 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
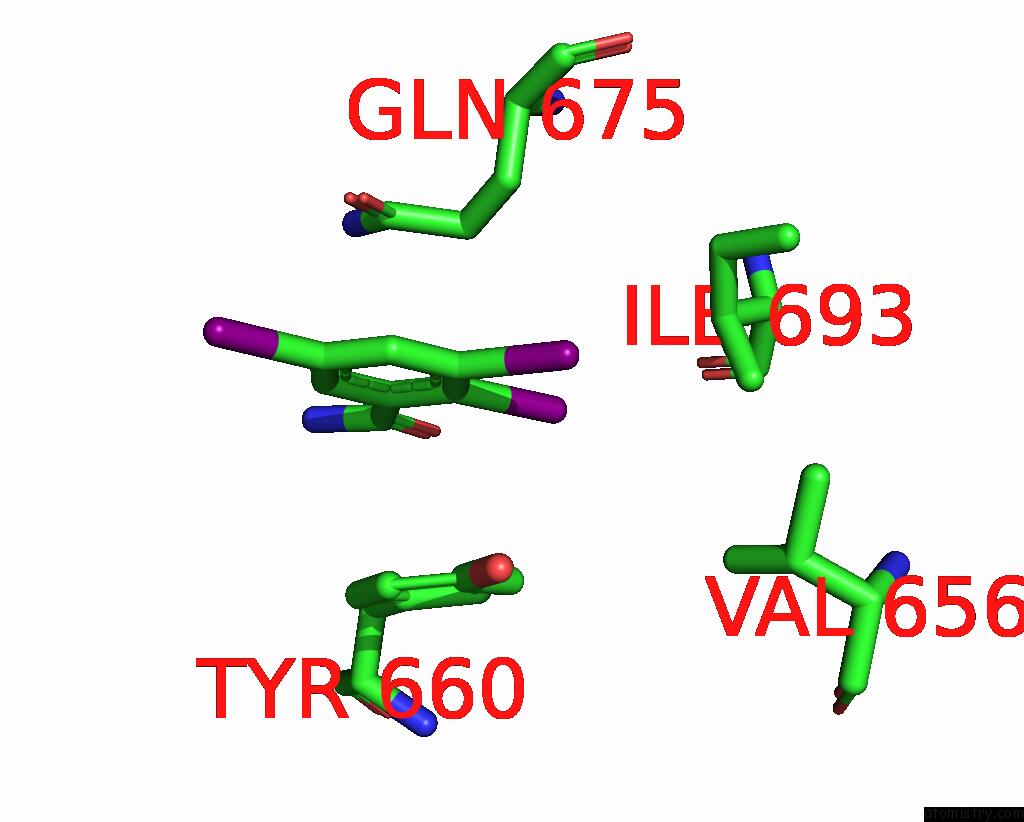
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 4 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
Iodine binding site 5 out of 9 in 7qur
Go back to
Iodine binding site 5 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 5 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
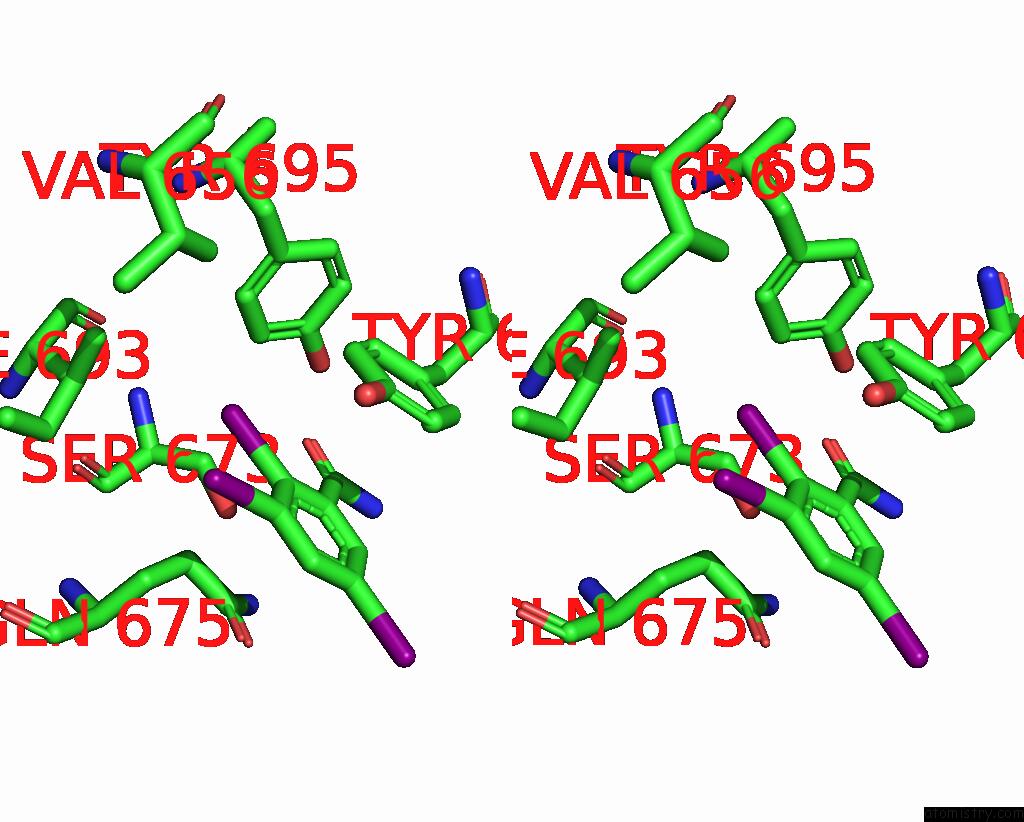
Iodine binding site 6 out of 9 in 7qur
Go back to
Iodine binding site 6 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
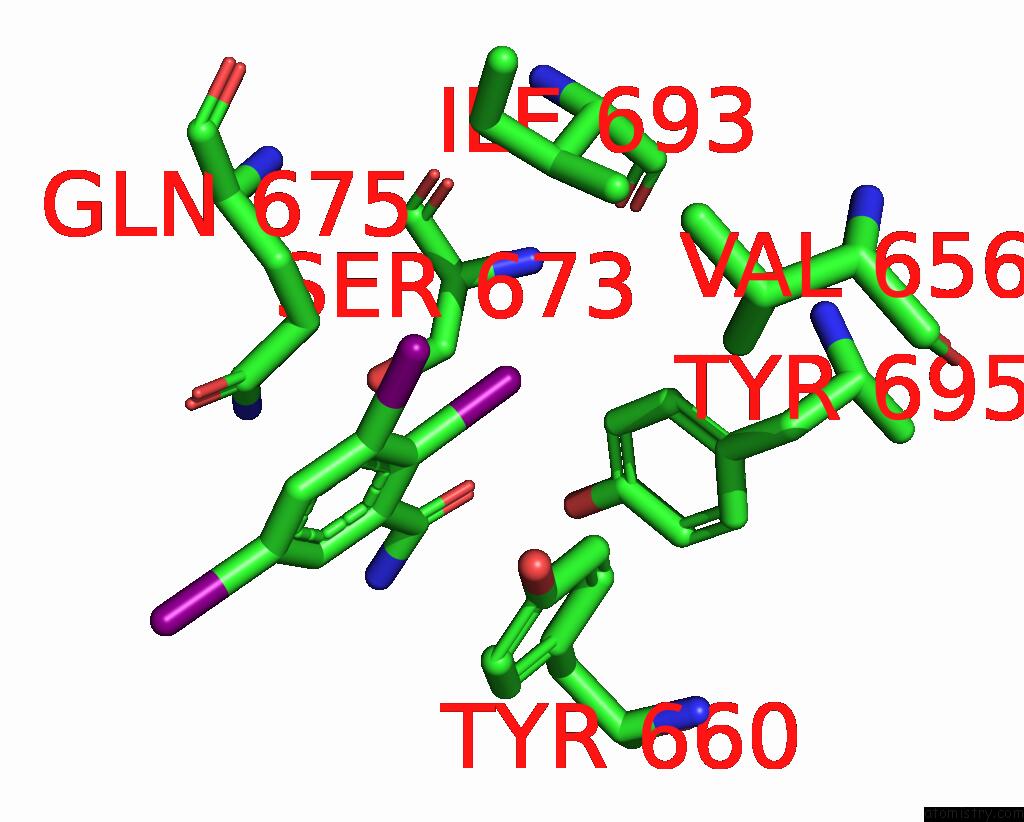
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 6 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
Iodine binding site 7 out of 9 in 7qur
Go back to
Iodine binding site 7 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 7 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
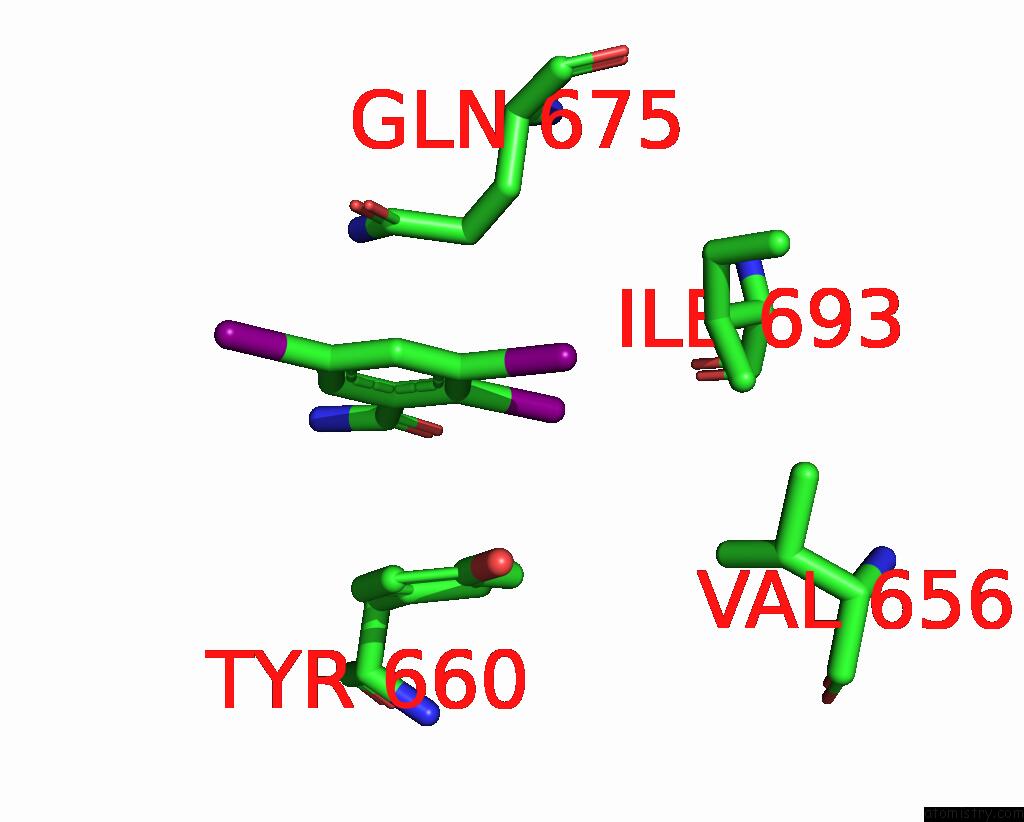
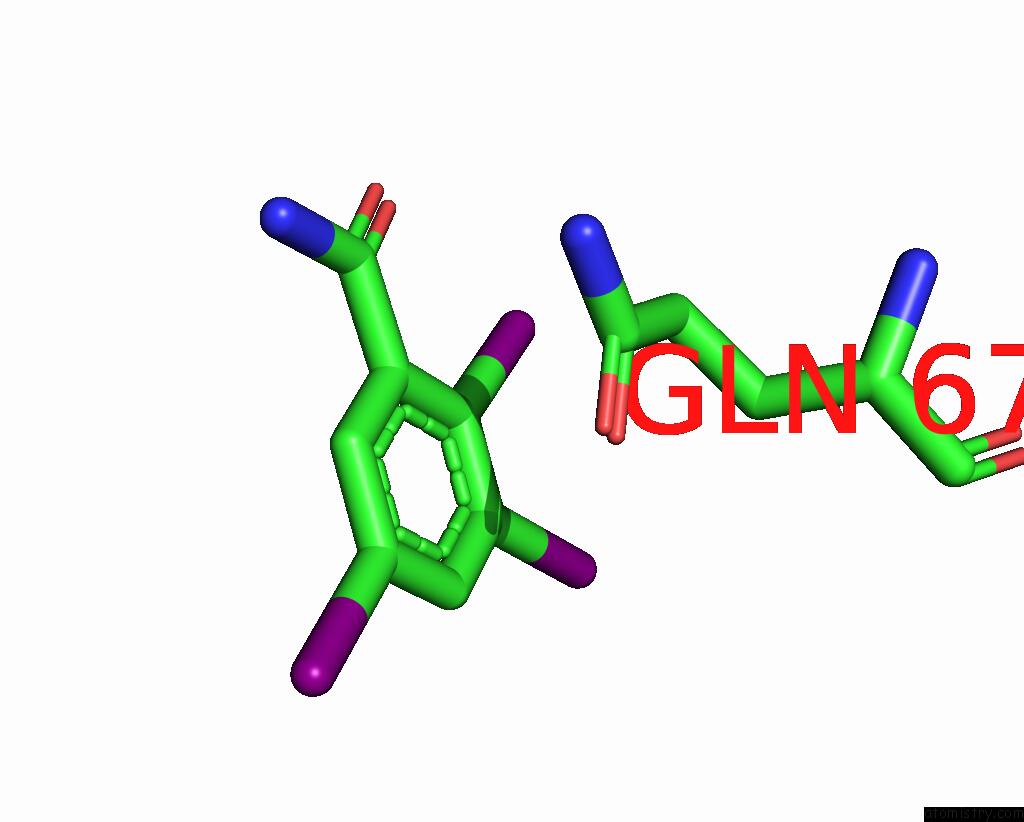
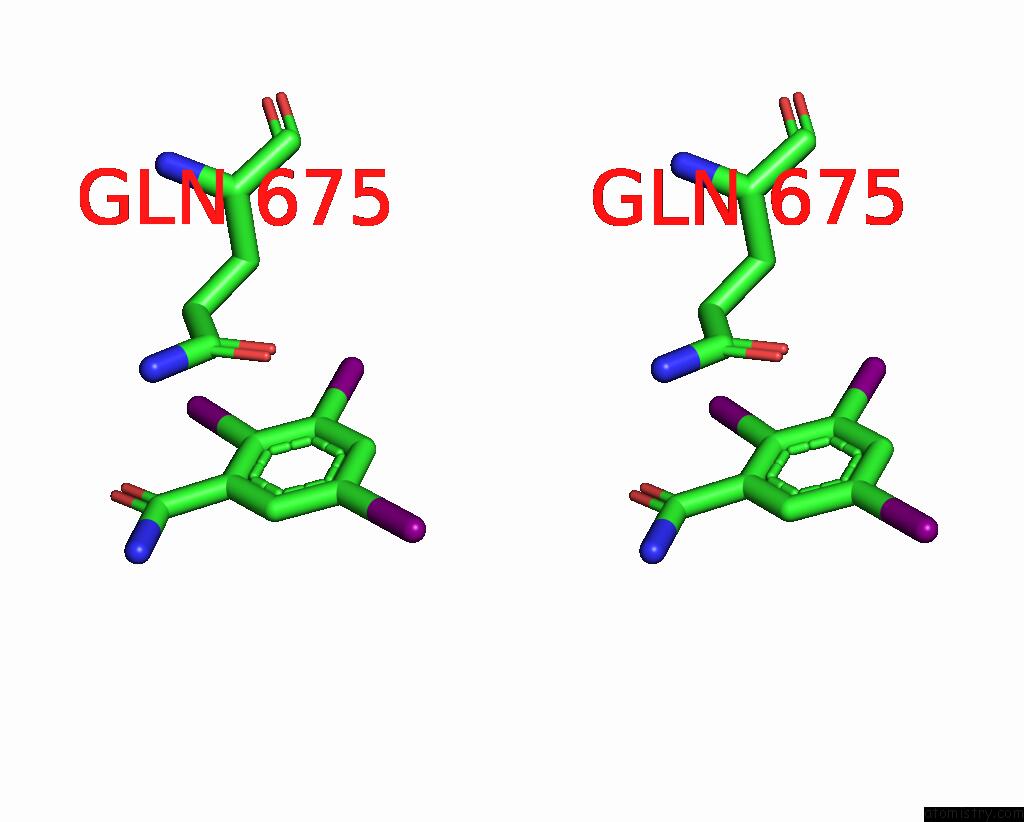
Iodine binding site 8 out of 9 in 7qur
Go back to
Iodine binding site 8 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 8 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
Iodine binding site 9 out of 9 in 7qur
Go back to
Iodine binding site 9 out
of 9 in the Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iodine with other atoms in the I binding
site number 9 of Sars-Cov-2 Spike with Ethylbenzamide-Tri-Iodo Siallyllactose, C3 Symmetry within 5.0Å range:
|
Reference:
C.J.Buchanan,
B.Gaunt,
P.J.Harrison,
Y.Yang,
J.Liu,
A.Khan,
A.M.Giltrap,
A.Le Bas,
P.N.Ward,
K.Gupta,
M.Dumoux,
T.K.Tan,
L.Schimaski,
S.Daga,
N.Picchiotti,
M.Baldassarri,
E.Benetti,
C.Fallerini,
F.Fava,
A.Giliberti,
P.I.Koukos,
M.J.Davy,
A.Lakshminarayanan,
X.Xue,
G.Papadakis,
L.P.Deimel,
V.Casablancas-Antras,
T.D.W.Claridge,
A.M.J.J.Bonvin,
Q.J.Sattentau,
S.Furini,
M.Gori,
J.Huo,
R.J.Owens,
C.Schaffitzel,
I.Berger,
A.Renieri,
J.H.Naismith,
A.J.Baldwin,
B.G.Davis.
Pathogen-Sugar Interactions Revealed By Universal Saturation Transfer Analysis. Science V. 377 M3125 2022.
ISSN: ESSN 1095-9203
PubMed: 35737812
DOI: 10.1126/SCIENCE.ABM3125
Page generated: Mon Aug 12 01:59:18 2024
ISSN: ESSN 1095-9203
PubMed: 35737812
DOI: 10.1126/SCIENCE.ABM3125
Last articles
I in 1GWDI in 1GUL
I in 1GTE
I in 1GTH
I in 1GJD
I in 1F3M
I in 1FZ9
I in 1GA5
I in 1F86
I in 1GA4